Construction of Circular RNA-MicroRNA-Messenger RNA Regulatory Network of Recurrent Implantation Failure to Explore Its Potential Pathogenesis
- PMID: 33664765
- PMCID: PMC7924221
- DOI: 10.3389/fgene.2020.627459
Construction of Circular RNA-MicroRNA-Messenger RNA Regulatory Network of Recurrent Implantation Failure to Explore Its Potential Pathogenesis
Abstract
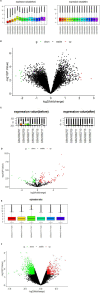
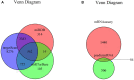
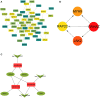
Background: Many studies on circular RNAs (circRNAs) have recently been published. However, the function of circRNAs in recurrent implantation failure (RIF) is unknown and remains to be explored. This study aims to determine the regulatory mechanisms of circRNAs in RIF. Methods: Microarray data of RIF circRNA (GSE147442), microRNA (miRNA; GSE71332), and messenger RNA (mRNA; GSE103465) were downloaded from the Gene Expression Omnibus (GEO) database to identify differentially expressed circRNA, miRNA, and mRNA. The circRNA-miRNA-mRNA network was constructed by Cytoscape 3.8.0 software, then the protein-protein interaction (PPI) network was constructed by STRING database, and the hub genes were identified by cytoHubba plug-in. The circRNA-miRNA-hub gene regulatory subnetwork was formed to understand the regulatory axis of hub genes in RIF. Finally, the Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the hub genes were performed by clusterProfiler package of Rstudio software, and Reactome Functional Interaction (FI) plug-in was used for reactome analysis to comprehensively analyze the mechanism of hub genes in RIF. Results: A total of eight upregulated differentially expressed circRNAs (DECs), five downregulated DECs, 56 downregulated differentially expressed miRNAs (DEmiRs), 104 upregulated DEmiRs, 429 upregulated differentially expressed genes (DEGs), and 1,067 downregulated DEGs were identified regarding RIF. The miRNA response elements of 13 DECs were then predicted. Seven overlapping miRNAs were obtained by intersecting the predicted miRNA and DEmiRs. Then, 56 overlapping mRNAs were obtained by intersecting the predicted target mRNAs of seven miRNAs with 1,496 DEGs. The circRNA-miRNA-mRNA network and PPI network were constructed through six circRNAs, seven miRNAs, and 56 mRNAs; and four hub genes (YWHAZ, JAK2, MYH9, and RAP2C) were identified. The circRNA-miRNA-hub gene regulatory subnetwork with nine regulatory axes was formed in RIF. Functional enrichment analysis and reactome analysis showed that these four hub genes were closely related to the biological functions and pathways of RIF. Conclusion: The results of this study provide further understanding of the potential pathogenesis from the perspective of circRNA-related competitive endogenous RNA network in RIF.
Keywords: GEO; circRNA; competitive endogenous RNA; network; recurrent implantation failure.
Copyright © 2021 Luo, Zhu, Zhou, Zhang, Zhang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Bellati F., Costanzi F., De Marco M. P., Cippitelli C., Stoppacciaro A., De Angelis C., et al. (2019). Low endometrial beta-catenin and cadherins expression patterns are predictive for primary infertility and recurrent pregnancy loss. Gynecol. Endocrinol. 35, 727–731. 10.1080/09513590.2019.1579790 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

