Identification of Predictor Genes for Feed Efficiency in Beef Cattle by Applying Machine Learning Methods to Multi-Tissue Transcriptome Data
- PMID: 33664767
- PMCID: PMC7921797
- DOI: 10.3389/fgene.2021.619857
Identification of Predictor Genes for Feed Efficiency in Beef Cattle by Applying Machine Learning Methods to Multi-Tissue Transcriptome Data
Abstract
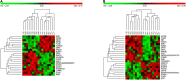
Machine learning (ML) methods have shown promising results in identifying genes when applied to large transcriptome datasets. However, no attempt has been made to compare the performance of combining different ML methods together in the prediction of high feed efficiency (HFE) and low feed efficiency (LFE) animals. In this study, using RNA sequencing data of five tissues (adrenal gland, hypothalamus, liver, skeletal muscle, and pituitary) from nine HFE and nine LFE Nellore bulls, we evaluated the prediction accuracies of five analytical methods in classifying FE animals. These included two conventional methods for differential gene expression (DGE) analysis (t-test and edgeR) as benchmarks, and three ML methods: Random Forests (RFs), Extreme Gradient Boosting (XGBoost), and combination of both RF and XGBoost (RX). Utility of a subset of candidate genes selected from each method for classification of FE animals was assessed by support vector machine (SVM). Among all methods, the smallest subsets of genes (117) identified by RX outperformed those chosen by t-test, edgeR, RF, or XGBoost in classification accuracy of animals. Gene co-expression network analysis confirmed the interactivity existing among these genes and their relevance within the network related to their prediction ranking based on ML. The results demonstrate a great potential for applying a combination of ML methods to large transcriptome datasets to identify biologically important genes for accurately classifying FE animals.
Keywords: Bos indicus; Extreme Gradient Boosting; RNA-seq; Random Forest; co-expression network; residual feed intake; supporting vector machine.
Copyright © 2021 Chen, Alexandre, Ribeiro, Fukumasu, Sun, Reverter and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Abo-Ismail M. K., Kelly M. J., Squires E. J., Swanson K. C., Bauck S., Miller S. P. (2013). Identification of single nucleotide polymorphisms in genes involved in digestive and metabolic processes associated with feed efficiency and performance traits in beef cattle. J. Anim. Sci. 91 2512–2529. 10.2527/jas.2012-5756 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

