Comparative transcriptomics of ice-crawlers demonstrates cold specialization constrains niche evolution in a relict lineage
- PMID: 33664782
- PMCID: PMC7896716
- DOI: 10.1111/eva.13120
Comparative transcriptomics of ice-crawlers demonstrates cold specialization constrains niche evolution in a relict lineage
Abstract
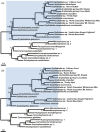
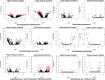
Key changes in ecological niche space are often critical to understanding how lineages diversify during adaptive radiations. However, the converse, or understanding why some lineages are depauperate and relictual, is more challenging, as many factors may constrain niche evolution. In the case of the insect order Grylloblattodea, highly conserved thermal breadth is assumed to be closely tied to their relictual status, but has not been formerly tested. Here, we investigate whether evolutionary constraints in the physiological tolerance of temperature can help explain relictualism in this lineage. Using a comparative transcriptomics approach, we investigate gene expression following acute heat and cold stress across members of Grylloblattodea and their sister group, Mantophasmatodea. We additionally examine patterns of protein evolution, to identify candidate genes of positive selection. We demonstrate that cold specialization in Grylloblattodea has been accompanied by the loss of the inducible heat shock response under both acute heat and cold stress. Additionally, there is widespread evidence of selection on protein-coding genes consistent with evolutionary constraints due to cold specialization. This includes positive selection on genes involved in trehalose transport, metabolic function, mitochondrial function, oxygen reduction, oxidative stress, and protein synthesis. These patterns of molecular adaptation suggest that Grylloblattodea have undergone evolutionary trade-offs to survive in cold habitats and should be considered highly vulnerable to climate change. Finally, our transcriptomic data provide a robust backbone phylogeny for generic relationships within Grylloblattodea and Mantophasmatodea. Major phylogenetic splits in each group relate to arid conditions driving biogeographical patterns, with support for a sister-group relationship between North American Grylloblatta and Altai-Sayan Grylloblattella, and a range disjunction in Namibia splitting major clades within Mantophasmatodea.
Keywords: Grylloblattodea; adaptation; gene regulation; insect phylogenomics; niche conservatism; protein evolution.
© 2020 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Figures


Similar articles
-
Conserved and narrow temperature limits in alpine insects: Thermal tolerance and supercooling points of the ice-crawlers, Grylloblatta (Insecta: Grylloblattodea: Grylloblattidae).J Insect Physiol. 2015 Jul;78:55-61. doi: 10.1016/j.jinsphys.2015.04.014. Epub 2015 May 5. J Insect Physiol. 2015. PMID: 25956197
-
Phylogeny and biogeography of ice crawlers (Insecta: Grylloblattodea) based on six molecular loci: designating conservation status for Grylloblattodea species.Mol Phylogenet Evol. 2006 Oct;41(1):222-37. doi: 10.1016/j.ympev.2006.04.013. Epub 2006 Apr 28. Mol Phylogenet Evol. 2006. PMID: 16798019
-
Evolutionary diversification of cryophilic Grylloblatta species (Grylloblattodea: Grylloblattidae) in alpine habitats of California.BMC Evol Biol. 2010 Jun 2;10:163. doi: 10.1186/1471-2148-10-163. BMC Evol Biol. 2010. PMID: 20525203 Free PMC article.
-
Heat freezes niche evolution.Ecol Lett. 2013 Sep;16(9):1206-19. doi: 10.1111/ele.12155. Epub 2013 Jul 22. Ecol Lett. 2013. PMID: 23869696 Review.
-
Insect responses to heat: physiological mechanisms, evolution and ecological implications in a warming world.Biol Rev Camb Philos Soc. 2020 Jun;95(3):802-821. doi: 10.1111/brv.12588. Epub 2020 Feb 8. Biol Rev Camb Philos Soc. 2020. PMID: 32035015 Review.
Cited by
-
Genetics of adaptation.Proc Natl Acad Sci U S A. 2022 Jul 26;119(30):e2122152119. doi: 10.1073/pnas.2122152119. Epub 2022 Jul 18. Proc Natl Acad Sci U S A. 2022. PMID: 35858399 Free PMC article.
-
Descending from trees: a Cretaceous winged ice-crawler illuminates the ecological shift and origin of Grylloblattidae.Proc Biol Sci. 2025 Jun;292(2049):20250557. doi: 10.1098/rspb.2025.0557. Epub 2025 Jun 18. Proc Biol Sci. 2025. PMID: 40527457 Free PMC article.
References
-
- Angilletta, Jr., M. J. , Wilson, R. S. , Navas, C. A. , & James, R. S. (2003). Tradeoffs and the evolution of thermal reaction norms. Trends in Ecology & Evolution, 18(5), 234–240. 10.1016/S0169-5347(03)00087-9 - DOI
LinkOut - more resources
Full Text Sources

