Population genomics and history of speciation reveal fishery management gaps in two related redfish species (Sebastes mentella and Sebastes fasciatus)
- PMID: 33664797
- PMCID: PMC7896722
- DOI: 10.1111/eva.13143
Population genomics and history of speciation reveal fishery management gaps in two related redfish species (Sebastes mentella and Sebastes fasciatus)
Abstract
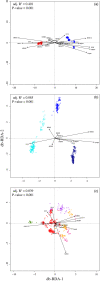
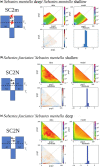
Understanding the processes shaping population structure and reproductive isolation of marine organisms can improve their management and conservation. Using genomic markers combined with estimation of individual ancestries, assignment tests, spatial ecology, and demographic modeling, we (i) characterized the contemporary population structure, (ii) assessed the influence of space, fishing depth, and sampling years on contemporary distribution, and (iii) reconstructed the speciation history of two cryptic redfish species, Sebastes mentella and S. fasciatus. We genotyped 860 individuals in the Northwest Atlantic Ocean using 24,603 filtered single nucleotide polymorphisms (SNPs). Our results confirmed the clear genetic distinctiveness of the two species and identified three ecotypes within S. mentella and five populations in S. fasciatus. Multivariate analyses highlighted the influence of spatial distribution and depth on the overall genomic variation, while demographic modeling revealed that secondary contact models best explained inter- and intragenomic divergence. These species, ecotypes, and populations can be considered as a rare and wide continuum of genomic divergence in the marine environment. This acquired knowledge pertaining to the evolutionary processes driving population divergence and reproductive isolation will help optimizing the assessment of demographic units and possibly to refine fishery management units.
Keywords: Sebastes; demographic models; fishery management; population genomics; related species; spatial ecology.
© 2020 Her Majesty the Queen in Right of Canada. Evolutionary Applications published by John Wiley & Sons Ltd. Reproduced with the permission of the Minister of Fishers and Ocean.
Figures




References
-
- Atkinson, D. B. (1987). The redfish resources off Canada’s east coast, pp. 15–33. In Proceedings of the Lowel Wakefield Fisheries Symposium: International Rockfish Symposium, Anchorage, AK, Alaska Sea grant College program report 97‐2.
LinkOut - more resources
Full Text Sources
Other Literature Sources

