Overexpression of EZH2/NSD2 Histone Methyltransferase Axis Predicts Poor Prognosis and Accelerates Tumor Progression in Triple-Negative Breast Cancer
- PMID: 33665162
- PMCID: PMC7921704
- DOI: 10.3389/fonc.2020.600514
Overexpression of EZH2/NSD2 Histone Methyltransferase Axis Predicts Poor Prognosis and Accelerates Tumor Progression in Triple-Negative Breast Cancer
Abstract
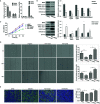
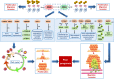
Two histone methyltransferases, enhancer of zeste homolog 2 (EZH2) and nuclear SET domain-containing 2 (NSD2), are aberrantly expressed in several types of human cancers. However, the regulatory relationship between EZH2 and NSD2 and their prognostic values in breast cancer (BC) have not been fully elucidated. In this study, we demonstrated that EZH2 and NSD2 were overexpressed in BC compared with benign lesions and normal tissues using tissue microarray, immunohistochemistry, and bioinformatic databases. Both EZH2 and NSD2 expression were associated with pathological grade of tumor and lymph node metastasis. A comprehensive survival analysis using Kaplan-Meier Plotter database indicated that EZH2 expression was negatively correlated with relapse-free survival (RFS), overall survival (OS), distant metastasis-free survival (DMFS), and postprogression survival (PPS) in 3951 BC patients, and NSD2 expression was negatively correlated with RFS and DMFS. Notably, EZH2 and NSD2 expression were coordinately higher in triple-negative breast cancer (TNBC) than that in other subtypes. Stable knockdown of EZH2 using lentiviral shRNA vector significantly reduced the proliferation, migration and invasion abilities of TNBC cell line MDA-MB-231 and MDA-MB-468, and downregulated NSD2 expression as well as the levels of H3K27me3 and H3K36me2, two histone methylation markers catalyzed by EZH2 and NSD2, respectively. By contrast, overexpression of EZH2 using adenovirus vector displayed an inverse phenotype. Furthermore, knockdown of NSD2 in EZH2-overexpressing cells could dramatically attenuate EZH2-mediated oncogenic effects. Bioinformatic analysis further revealed the function and pathway enrichments of co-expressed genes and interactive genes of EZH2/NSD2 axis, suggesting that EZH2/NSD2 axis was associated with cell division, mitotic nuclear division and transition of mitotic cell cycle in TNBC. Taken together, EZH2/NSD2 axis may act as a predictive marker for poor prognosis and accelerate the progression of TNBC.
Keywords: EZH2; NSD2; prognosis; progression; triple-negative breast cancer.
Copyright © 2021 Gao, Liu, Li, Zhao and Pan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

