Zinc Finger Protein CTCF Regulates Extracellular Matrix (ECM)-Related Gene Expression Associated With the Wnt Signaling Pathway in Gastric Cancer
- PMID: 33665169
- PMCID: PMC7921701
- DOI: 10.3389/fonc.2020.625633
Zinc Finger Protein CTCF Regulates Extracellular Matrix (ECM)-Related Gene Expression Associated With the Wnt Signaling Pathway in Gastric Cancer
Abstract
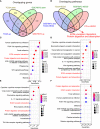
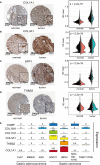
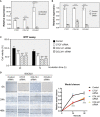
Gastric cancer (GC), a leading cause of cancer-related death, is a heterogeneous disease. We aim to describe clinically relevant molecular classifications of GC that incorporate heterogeneity and provide useful clinical information. We combined different gene expression datasets and filtered a 7-gene signature related to the extracellular matrix (ECM), which also exhibited significant prognostic value in GC patients. Interestingly, putative CCCTC-binding factor (CTCF) regulatory elements were identified within the promoters of these ECM-related genes and were confirmed by chromatin immunoprecipitation sequencing (ChIP-Seq). CTCF binding sites also overlapped with histone activation markers, indicating direct regulation. In addition, CTCF was also correlated with the Wnt signaling pathway. A comparison of human GC cell lines with high or low expression of ECM-related genes revealed different levels of tumor aggressiveness, suggesting the cancer development-promoting functions of ECM-related genes. Furthermore, CTCF regulated COL1A1 and COLA31 expression in vitro. Silencing CTCF or COL1A1/COL1A3 markedly inhibited cell growth and migration in the metastatic GC cell line BGC823. Collectively, this ECM-related 7-gene signature provides a novel insight for survival prediction among GC patients. The zinc finger protein CTCF regulates ECM-related genes, thereby promoting GC cell growth and migration.
Keywords: Wnt signaling; extracellular matrix (ECM); gastric cancer; histone modification; zinc finger protein CTCF.
Copyright © 2021 Liu, Deng, Lin, Zhang, Huang, Zhao, Jin, Xu, Ni, Xu, Ying and Hu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Exploring the changing landscape of cell-to-cell variation after CTCF knockdown via single cell RNA-seq.BMC Genomics. 2019 Dec 26;20(1):1015. doi: 10.1186/s12864-019-6379-5. BMC Genomics. 2019. PMID: 31878887 Free PMC article.
-
Exploring the Interaction between the SWI/SNF Chromatin Remodeling Complex and the Zinc Finger Factor CTCF.Int J Mol Sci. 2020 Nov 25;21(23):8950. doi: 10.3390/ijms21238950. Int J Mol Sci. 2020. PMID: 33255744 Free PMC article.
-
CTCF and CohesinSA-1 Mark Active Promoters and Boundaries of Repressive Chromatin Domains in Primary Human Erythroid Cells.PLoS One. 2016 May 24;11(5):e0155378. doi: 10.1371/journal.pone.0155378. eCollection 2016. PLoS One. 2016. PMID: 27219007 Free PMC article.
-
Modulation of thyroid hormone receptor silencing function by co-repressors and a synergizing transcription factor.Biochem Soc Trans. 2000;28(4):386-9. Biochem Soc Trans. 2000. PMID: 10961925 Review.
-
CTCF as a multifunctional protein in genome regulation and gene expression.Exp Mol Med. 2015 Jun 5;47(6):e166. doi: 10.1038/emm.2015.33. Exp Mol Med. 2015. PMID: 26045254 Free PMC article. Review.
Cited by
-
Gastrointestinal pan-cancer landscape of tumor matrix heterogeneity identifies biologically distinct matrix stiffness subtypes predicting prognosis and chemotherapy efficacy.Comput Struct Biotechnol J. 2023 Apr 20;21:2744-2758. doi: 10.1016/j.csbj.2023.04.016. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37181656 Free PMC article.
-
Potential Biomarkers and Signaling Pathways Associated with the Pathogenesis of Primary Ameloblastoma: A Systems Biology Approach.Int J Dent. 2022 Sep 16;2022:3316313. doi: 10.1155/2022/3316313. eCollection 2022. Int J Dent. 2022. PMID: 36160115 Free PMC article.
-
Integrated Bioinformatics and Experimental Approaches Identified the Role of NPPA in the Proliferation and the Malignant Behavior of Breast Cancer.J Immunol Res. 2021 Sep 27;2021:7876489. doi: 10.1155/2021/7876489. eCollection 2021. J Immunol Res. 2021. PMID: 34616853 Free PMC article.
-
Extracellular Matrix-Associated Pathways Promote the Progression of Gastric Cancer by Impacting the Dendritic Cell Axis.Int J Gen Med. 2021 Oct 13;14:6725-6739. doi: 10.2147/IJGM.S334245. eCollection 2021. Int J Gen Med. 2021. PMID: 34675633 Free PMC article.
-
Zinc Finger Proteins in the War on Gastric Cancer: Molecular Mechanism and Clinical Potential.Cells. 2023 May 4;12(9):1314. doi: 10.3390/cells12091314. Cells. 2023. PMID: 37174714 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

