Activation of amino acid metabolic program in cardiac HIF1-alpha-deficient mice
- PMID: 33665549
- PMCID: PMC7900219
- DOI: 10.1016/j.isci.2021.102124
Activation of amino acid metabolic program in cardiac HIF1-alpha-deficient mice
Abstract
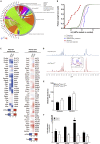
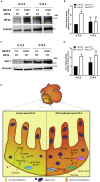
HIF1-alpha expression defines metabolic compartments in the developing heart, promoting glycolytic program in the compact myocardium and mitochondrial enrichment in the trabeculae. Nonetheless, its role in cardiogenesis is debated. To assess the importance of HIF1-alpha during heart development and the influence of glycolysis in ventricular chamber formation, herein we generated conditional knockout models of Hif1a in Nkx2.5 cardiac progenitors and cardiomyocytes. Deletion of Hif1a impairs embryonic glycolysis without influencing cardiomyocyte proliferation and results in increased mitochondrial number and transient activation of amino acid catabolism together with HIF2α and ATF4 upregulation by E12.5. Hif1a mutants display normal fatty acid oxidation program and do not show cardiac dysfunction in the adulthood. Our results demonstrate that cardiac HIF1 signaling and glycolysis are dispensable for mouse heart development and reveal the metabolic flexibility of the embryonic myocardium to consume amino acids, raising the potential use of alternative metabolic substrates as therapeutic interventions during ischemic events.
Keywords: Animal Physiology; Biological Sciences; Cellular Physiology; Developmental Biology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Averous J., Bruhat A., Jousse C., Carraro V., Thiel G., Fafournoux P. Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J. Biol. Chem. 2004;279:5288–5297. - PubMed
-
- Badiola N., Penas C., Miñano-Molina A., Barneda-Zahonero B., Fadó R., Sánchez-Opazo G., Comella J.X., Sabriá J., Zhu C., Blomgren K., et al. Induction of ER stress in response to oxygen-glucose deprivation of cortical cultures involves the activation of the PERK and IRE-1 pathways and of caspase-12. Cell Death Dis. 2011;2:e149. - PMC - PubMed
-
- Bing R.J., Siegel A., Ungar I., Gilbert M. Metabolism of the human heart: II. Studies on fat, ketone and amino acid metabolism. Am. J. Med. 1954;16:504–515. - PubMed
-
- Bohuslavova R., Skvorova L., Sedmera D., Semenza G.L., Pavlinkova G. Increased susceptibility of HIF-1α heterozygous-null mice to cardiovascular malformations associated with maternal diabetes. J. Mol. Cell. Cardiol. 2013;60:129–141. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

