Oral seeding and niche-adaptation of middle ear biofilms in health
- PMID: 33665609
- PMCID: PMC7822943
- DOI: 10.1016/j.bioflm.2020.100041
Oral seeding and niche-adaptation of middle ear biofilms in health
Abstract
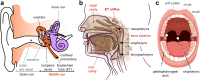
The entrenched dogma of a sterile middle ear mucosa in health is incongruent with its periodic aeration and seeding with saliva aerosols. To test this, we sequenced 16S rRNA-V4 amplicons from otic secretions collected at the nasopharyngeal orifice of the tympanic tube and, as controls, oropharyngeal and buccal samples. The otic samples harbored a rich diversity of oral keystone genera and similar functional traits but were enriched in anaerobic genera in the Bacteroidetes (Prevotella and Alloprevotella), Fusobacteria (Fusobacterium and Leptotrichia) and Firmicutes (Veillonella) phyla. Facultative anaerobes in the Streptococcus genus were also abundant in the otic and oral samples but corresponded to distinct, and sometimes novel, cultivars, consistent with the ecological diversification of the oral migrants once in the middle ear microenvironment. Neutral community models also predicted a large contribution of oral dispersal to the otic communities and the positive selection of taxa better adapted to growth and reproduction under limited aeration. These results challenge the traditional view of a sterile middle ear in health and highlight hitherto unknown roles for oral dispersal and episodic ventilation in seeding and diversifying otic biofilms.
Keywords: Divers; ET, Eustachian tube; Eustachian tube; Middle ear; OTUs, Operational taxonomic units; Oral dispersal; Otic infections; Otic microbiome; PCR, Polymerase chain reaction; PCoA, Principle coordinates analysis; rRNA, Ribosomal RNA.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Bassis C.M., Erb-Downward J.R., Dickson R.P., Freeman C.M., Schmidt T.M., Young V.B., Beck J.M., Curtis J.L., Huffnagle G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio. 2015;6 doi: 10.1128/mBio.00037-15. - DOI - PMC - PubMed
-
- Bluestone C.D. BC Decker Inc; Hamilton, Ontario: 2005. The Eustachian Tube: structure, function, and role in the middle ear.
-
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

