Immunization and Drug Metabolizing Enzymes: Focus on Hepatic Cytochrome P450 3A
- PMID: 33666138
- PMCID: PMC8292194
- DOI: 10.1080/14760584.2021.1899818
Immunization and Drug Metabolizing Enzymes: Focus on Hepatic Cytochrome P450 3A
Abstract
Objective: Infectious disease emergencies like the 2013-2016 Ebola epidemic and the 2009 influenza and current SARS-CoV-2 pandemics illustrate that vaccines are now given to diverse populations with preexisting pathologies requiring pharmacological management. Many natural biomolecules (steroid hormones, fatty acids, vitamins) and ~60% of prescribed medications are processed by hepatic cytochrome P450 (CYP) 3A4. The objective of this work was to determine the impact of infection and vaccines on drug metabolism.
Methods: The impact of an adenovirus-based vaccine expressing Ebola glycoprotein (AdEBO) and H1N1 and H3N2 influenza viruses on hepatic CYP 3A4 and associated nuclear receptors was evaluated in human hepatocytes (HC-04 cells) and in mice.
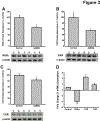
Results: CYP3A activity was suppressed by 55% in mice 24 h after administration of mouse-adapted H1N1, while ˂10% activity remained in HC-04 cells after infection with H1N1 and H3N2 due to global suppression of cellular translation capacity, indicated by reduction (70%, H1N1, 56%, H3N2) of phosphorylated eukaryotic translation initiation factor 4e (eIF4E). AdEBO suppressed CYP3A activity in vivo (44%) and in vitro (26%) 24 hours after infection.
Conclusion: As the clinical evaluation of vaccines for SARS-CoV-2 and other global pathogens rise, studies to evaluate the impact of new vaccines and emerging pathogens on CYP3A4 and other metabolic enzymes are warranted to avoid therapeutic failures that could further compromise the public health during infectious disease emergencies.
Keywords: Cytochrome P450 3A4; drug metabolism; ebola; influenza; liver; vaccine.
Conflict of interest statement
Figures
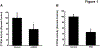


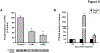
References
-
- Ortiz de Montellano PR (2019) The Cytochrome P450 Oxidative System In Pearson PG, Wienkers LC (Eds.), Handbook of Drug Metabolism, Third Edition, Boca Raton, FL, CRC Press, pp. 57–83.
-
- Coleman MD (2020) Drug Biotransformational Systems – Origins and Aims, In Human Drug Metabolism Third Edition, Hoboken, NJ, Wiley-Blackwell, pp.17–97.
-
- United States Food and Drug Administration, 2020, Drug Development and Drug Interactions: Table of Substrates, Inhibitors and Inducers, accessed 26 January 2021, <https://www.fda.gov/drugs/drug-interactions-labeling/drug-development-an...>
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
