Effective dynamics of nucleosome configurations at the yeast PHO5 promoter
- PMID: 33666171
- PMCID: PMC8004102
- DOI: 10.7554/eLife.58394
Effective dynamics of nucleosome configurations at the yeast PHO5 promoter
Abstract
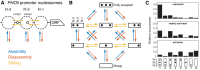
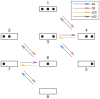
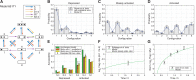
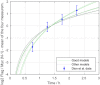
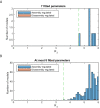
Chromatin dynamics are mediated by remodeling enzymes and play crucial roles in gene regulation, as established in a paradigmatic model, the Saccharomyces cerevisiae PHO5 promoter. However, effective nucleosome dynamics, that is, trajectories of promoter nucleosome configurations, remain elusive. Here, we infer such dynamics from the integration of published single-molecule data capturing multi-nucleosome configurations for repressed to fully active PHO5 promoter states with other existing histone turnover and new chromatin accessibility data. We devised and systematically investigated a new class of 'regulated on-off-slide' models simulating global and local nucleosome (dis)assembly and sliding. Only seven of 68,145 models agreed well with all data. All seven models involve sliding and the known central role of the N-2 nucleosome, but regulate promoter state transitions by modulating just one assembly rather than disassembly process. This is consistent with but challenges common interpretations of previous observations at the PHO5 promoter and suggests chromatin opening by binding competition.
Keywords: PHO5 promoter; S. cerevisiae; chromatin regulation; chromatin remodeling; chromosomes; computational biology; gene expression; nucleosome configurations; nucleosome dynamics; systems biology.
© 2021, Wolff et al.
Conflict of interest statement
MW, AS, PK, UG No competing interests declared
Figures













Similar articles
-
In vitro reconstitution of PHO5 promoter chromatin remodeling points to a role for activator-nucleosome competition in vivo.Mol Cell Biol. 2010 Aug;30(16):4060-76. doi: 10.1128/MCB.01399-09. Epub 2010 Jun 21. Mol Cell Biol. 2010. PMID: 20566699 Free PMC article.
-
Role of transcription factor-mediated nucleosome disassembly in PHO5 gene expression.Sci Rep. 2016 Feb 4;6:20319. doi: 10.1038/srep20319. Sci Rep. 2016. PMID: 26843321 Free PMC article.
-
In vivo role for the chromatin-remodeling enzyme SWI/SNF in the removal of promoter nucleosomes by disassembly rather than sliding.J Biol Chem. 2011 Nov 25;286(47):40556-65. doi: 10.1074/jbc.M111.289918. Epub 2011 Oct 6. J Biol Chem. 2011. PMID: 21979950 Free PMC article.
-
The yeast PHO5 promoter: from single locus to systems biology of a paradigm for gene regulation through chromatin.Nucleic Acids Res. 2014;42(17):10888-902. doi: 10.1093/nar/gku784. Epub 2014 Sep 4. Nucleic Acids Res. 2014. PMID: 25190457 Free PMC article. Review.
-
Yeast chromatin remodeling complexes and their roles in transcription.Curr Genet. 2020 Aug;66(4):657-670. doi: 10.1007/s00294-020-01072-0. Epub 2020 Apr 1. Curr Genet. 2020. PMID: 32239283 Review.
Cited by
-
Natural promoters and promoter engineering strategies for metabolic regulation in Saccharomyces cerevisiae.J Ind Microbiol Biotechnol. 2023 Feb 17;50(1):kuac029. doi: 10.1093/jimb/kuac029. J Ind Microbiol Biotechnol. 2023. PMID: 36633543 Free PMC article. Review.
-
Inositol pyrophosphate dynamics reveals control of the yeast phosphate starvation program through 1,5-IP8 and the SPX domain of Pho81.Elife. 2023 Sep 20;12:RP87956. doi: 10.7554/eLife.87956. Elife. 2023. PMID: 37728314 Free PMC article.
-
Single-Molecule Techniques to Study Chromatin.Front Cell Dev Biol. 2021 Jul 5;9:699771. doi: 10.3389/fcell.2021.699771. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34291054 Free PMC article. Review.
-
Allosteric conformational ensembles have unlimited capacity for integrating information.Elife. 2021 Jun 9;10:e65498. doi: 10.7554/eLife.65498. Elife. 2021. PMID: 34106049 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

