RNA helicase DDX5 acts as a critical regulator for survival of neonatal mouse gonocytes
- PMID: 33666296
- PMCID: PMC8088469
- DOI: 10.1111/cpr.13000
RNA helicase DDX5 acts as a critical regulator for survival of neonatal mouse gonocytes
Abstract
Objectives: Mammalian spermatogenesis is a biological process of male gamete formation. Gonocytes are the only precursors of spermatogonial stem cells (SSCs) which develop into mature spermatozoa. DDX5 is one of DEAD-box RNA helicases and expresses in male germ cells, suggesting that Ddx5 plays important functions during spermatogenesis. Here, we explore the functions of Ddx5 in regulating the specification of gonocytes.
Materials and methods: Germ cell-specific Ddx5 knockout (Ddx5-/- ) mice were generated. The morphology of testes and epididymides and fertility in both wild-type and Ddx5-/- mice were analysed. Single-cell RNA sequencing (scRNA-seq) was used to profile the transcriptome in testes from wild-type and Ddx5-/- mice at postnatal day (P) 2. Dysregulated genes were validated by single-cell qRT-PCR and immunofluorescent staining.
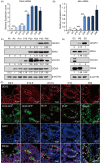
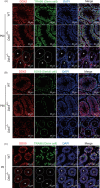
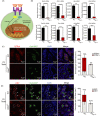
Results: In male mice, Ddx5 was expressed in germ cells at different stages of development. Germ cell-specific Ddx5 knockout adult male mice were sterile due to completely devoid of germ cells. Male germ cells gradually disappeared in Ddx5-/- mice from E18.5 to P6. Single-cell transcriptome analysis showed that genes involved in cell cycle and glial cell line-derived neurotrophic factor (GDNF) pathway were significantly decreased in Ddx5-deficient gonocytes. Notably, Ddx5 ablation impeded the proliferation of gonocytes.
Conclusions: Our study reveals the critical roles of Ddx5 in fate determination of gonocytes, offering a novel insight into the pathogenesis of male sterility.
Keywords: DDX5; RNA-binding protein; gonocyte; spermatogonial stem cell; testis.
© 2021 The Authors. Cell Proliferation Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293‐300. - PubMed
-
- Saitou M. Specification of the germ cell lineage in mice. Front Biosci (Landmark Ed). 2009;14:1068‐1087. - PubMed
-
- Anderson R, Copeland TK, Scholer H, et al. The onset of germ cell migration in the mouse embryo. Mech Dev. 2000;91:61‐68. - PubMed
-
- Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521‐528. - PubMed
MeSH terms
Substances
Grants and funding
- 2016YFA0100300/The Ministry of Science and Technology of the People's Republic of China
- XDA16010502/Strategic Priority Research Program of the Chinese Academy of Sciences
- XDA16020404/Strategic Priority Research Program of the Chinese Academy of Sciences
- 2016YFA0100400/The Ministry of Science and Technology of the People's Republic of China
- 31925009/National Natural Science Foundation of China
- 81902885/National Natural Science Foundation of China
- 32000424/National Natural Science Foundation of China
- 31871456/National Natural Science Foundation of China
- 2018GZR110104007/Key Research & Development Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory
- 2019B020234004/Science and Technology Planning Project of Guangdong Province, China
- 2020B1212060052/Science and Technology Planning Project of Guangdong Province, China
- 201906010096/Science and Technology Program of Guangzhou, China
- 2019A1515110028/Guangdong Basic and Applied Basic Research Foundation
- 2019B151502054/Guangdong Basic and Applied Basic Research Foundation
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

