Amyloid burden quantification depends on PET and MR image processing methodology
- PMID: 33667281
- PMCID: PMC7935288
- DOI: 10.1371/journal.pone.0248122
Amyloid burden quantification depends on PET and MR image processing methodology
Abstract
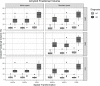
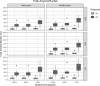
Quantification of amyloid load with positron emission tomography can be useful to assess Alzheimer's Disease in-vivo. However, quantification can be affected by the image processing methodology applied. This study's goal was to address how amyloid quantification is influenced by different semi-automatic image processing pipelines. Images were analysed in their Native Space and Standard Space; non-rigid spatial transformation methods based on maximum a posteriori approaches and tissue probability maps (TPM) for regularisation were explored. Furthermore, grey matter tissue segmentations were defined before and after spatial normalisation, and also using a population-based template. Five quantification metrics were analysed: two intensity-based, two volumetric-based, and one multi-parametric feature. Intensity-related metrics were not substantially affected by spatial normalisation and did not significantly depend on the grey matter segmentation method, with an impact similar to that expected from test-retest studies (≤10%). Yet, volumetric and multi-parametric features were sensitive to the image processing methodology, with an overall variability up to 45%. Therefore, the analysis should be carried out in Native Space avoiding non-rigid spatial transformations. For analyses in Standard Space, spatial normalisation regularised by TPM is preferred. Volumetric-based measurements should be done in Native Space, while intensity-based metrics are more robust against differences in image processing pipelines.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group* under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34: 939–939. 10.1212/wnl.34.7.939 - DOI - PubMed
-
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Kawas CH, et al.. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7: 263–269. 10.1016/j.jalz.2011.03.005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

