Type I collagen deletion in αSMA+ myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer
- PMID: 33667385
- PMCID: PMC8423173
- DOI: 10.1016/j.ccell.2021.02.007
Type I collagen deletion in αSMA+ myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer
Abstract
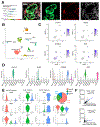
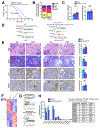
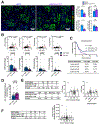
Stromal desmoplastic reaction in pancreatic ductal adenocarcinoma (PDAC) involves significant accumulation of type I collagen (Col1). However, the precise molecular and mechanistic contribution of Col1 in PDAC progression remains unknown. Activated pancreatic stellate cells/αSMA+ myofibroblasts are major contributors of Col1 in the PDAC stroma. We use a dual-recombinase genetic mouse model of spontaneous PDAC to delete Col1 specifically in myofibroblasts. This results in significant reduction of total stromal Col1 content and accelerates the emergence of PanINs and PDAC, decreasing overall survival. Col1 deletion leads to Cxcl5 upregulation in cancer cells via SOX9. Increase in Cxcl5 is associated with recruitment of myeloid-derived suppressor cells and suppression of CD8+ T cells, which can be attenuated with combined targeting of CXCR2 and CCR2 to restrain accelerated PDAC progression in the setting of stromal Col1 deletion. Our results unravel the fundamental role of myofibroblast-derived Co1l in regulating tumor immunity and restraining PDAC progression.
Keywords: B cells; T cells; extracellular matrix; fibroblasts; genetically engineered mouse models; myeloid-derived suppressor cells (MDSCs); pancreatic ductal adenocarcinoma (PDAC); tumor immunology; tumor microenvironment; type I collagen.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no conflict of interest.
Figures





Comment in
-
Pancreatic cancer is suppressed by fibroblast-derived collagen I.Cancer Cell. 2021 Apr 12;39(4):451-453. doi: 10.1016/j.ccell.2021.02.017. Epub 2021 Mar 11. Cancer Cell. 2021. PMID: 33711271
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

