The human pandemic coronaviruses on the show: The spike glycoprotein as the main actor in the coronaviruses play
- PMID: 33667553
- PMCID: PMC7921731
- DOI: 10.1016/j.ijbiomac.2021.02.203
The human pandemic coronaviruses on the show: The spike glycoprotein as the main actor in the coronaviruses play
Abstract
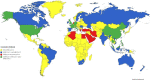
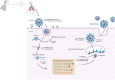
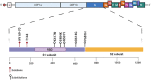
Three coronaviruses (CoVs) have threatened the world population by causing outbreaks in the last two decades. In late 2019, the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) emerged and caused the coronaviruses to disease 2019 (COVID-19), leading to the ongoing global outbreak. The other pandemic coronaviruses, SARS-CoV and Middle East respiratory syndrome CoV (MERS-CoV), share a considerable level of similarities at genomic and protein levels. However, the differences between them lead to distinct behaviors. These differences result from the accumulation of mutations in the sequence and structure of spike (S) glycoprotein, which plays an essential role in coronavirus infection, pathogenicity, transmission, and evolution. In this review, we brought together many studies narrating a sequence of events and highlighting the differences among S proteins from SARS-CoV, MERS-CoV, and SARS-CoV-2. It was performed here, analysis of S protein sequences and structures from the three pandemic coronaviruses pointing out the mutations among them and what they come through. Additionally, we investigated the receptor-binding domain (RBD) from all S proteins explaining the mutation and biological importance of all of them. Finally, we discuss the mutation in the S protein from several new isolates of SARS-CoV-2, reporting their difference and importance. This review brings into detail how the variations in S protein that make SARS-CoV-2 more aggressive than its relatives coronaviruses and other differences between coronaviruses.
Keywords: Coronaviruses; MERS-CoV; Mutations; RBD; SARS-CoV; SARS-CoV-2; Spike proteins.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest All authors declare no conflicts of interest.
Figures




References
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. - PMC - PubMed
-
- ICTV https://talk.ictvonline.org/taxonomy/ Available online: (accessed on Jan 21, 2021)
-
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/nejmoa030747. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

