Genomic Analysis, Progress and Future Perspectives in Dairy Cattle Selection: A Review
- PMID: 33668747
- PMCID: PMC7996307
- DOI: 10.3390/ani11030599
Genomic Analysis, Progress and Future Perspectives in Dairy Cattle Selection: A Review
Abstract
Genomics comprises a set of current and valuable technologies implemented as selection tools in dairy cattle commercial breeding programs. The intensive progeny testing for production and reproductive traits based on genomic breeding values (GEBVs) has been crucial to increasing dairy cattle productivity. The knowledge of key genes and haplotypes, including their regulation mechanisms, as markers for productivity traits, may improve the strategies on the present and future for dairy cattle selection. Genome-wide association studies (GWAS) such as quantitative trait loci (QTL), single nucleotide polymorphisms (SNPs), or single-step genomic best linear unbiased prediction (ssGBLUP) methods have already been included in global dairy programs for the estimation of marker-assisted selection-derived effects. The increase in genetic progress based on genomic predicting accuracy has also contributed to the understanding of genetic effects in dairy cattle offspring. However, the crossing within inbred-lines critically increased homozygosis with accumulated negative effects of inbreeding like a decline in reproductive performance. Thus, inaccurate-biased estimations based on empirical-conventional models of dairy production systems face an increased risk of providing suboptimal results derived from errors in the selection of candidates of high genetic merit-based just on low-heritability phenotypic traits. This extends the generation intervals and increases costs due to the significant reduction of genetic gains. The remarkable progress of genomic prediction increases the accurate selection of superior candidates. The scope of the present review is to summarize and discuss the advances and challenges of genomic tools for dairy cattle selection for optimizing breeding programs and controlling negative inbreeding depression effects on productivity and consequently, achieving economic-effective advances in food production efficiency. Particular attention is given to the potential genomic selection-derived results to facilitate precision management on modern dairy farms, including an overview of novel genome editing methodologies as perspectives toward the future.
Keywords: dairy cattle; environment; gene edition; genomic analysis; health; linear types; nutrition; production; reproduction; welfare.
Conflict of interest statement
The authors participating in this review declare not to have conflicts of interest.
Figures
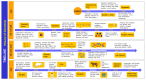
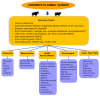
References
Publication types
Grants and funding
- 20/10/UTC2020/Universidad Técnica de Cotopaxi (UTC), Ecuador.
- 2020 - 21201280/Universidad San Francisco de Quito (USFQ), Ecuador; Secretariat of Higher Education, Science, Technology and Innovation (SENESCYT) of the Government of Ecuador; Agencia Nacional de Investigación y Desarrollo (ANID); Programa de Becas / Doctorado Nacional
LinkOut - more resources
Full Text Sources
Other Literature Sources

