Probing the Role of the Conserved Arg174 in Formate Dehydrogenase by Chemical Modification and Site-Directed Mutagenesis
- PMID: 33668802
- PMCID: PMC7956174
- DOI: 10.3390/molecules26051222
Probing the Role of the Conserved Arg174 in Formate Dehydrogenase by Chemical Modification and Site-Directed Mutagenesis
Abstract
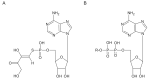
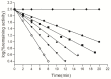
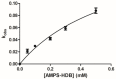
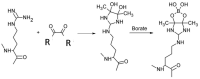
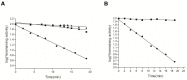
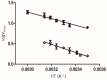
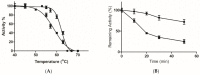
The reactive adenosine derivative, adenosine 5'-O-[S-(4-hydroxy-2,3-dioxobutyl)]-thiophosphate (AMPS-HDB), contains a dicarbonyl group linked to the purine nucleotide at a position equivalent to the pyrophosphate region of NAD+. AMPS-HDB was used as a chemical label towards Candida boidinii formate dehydrogenase (CbFDH). AMPS-HDB reacts covalently with CbFDH, leading to complete inactivation of the enzyme activity. The inactivation kinetics of CbFDH fit the Kitz and Wilson model for time-dependent, irreversible inhibition (KD = 0.66 ± 0.15 mM, first order maximum rate constant k3 = 0.198 ± 0.06 min-1). NAD+ and NADH protects CbFDH from inactivation by AMPS-HDB, showing the specificity of the reaction. Molecular modelling studies revealed Arg174 as a candidate residue able to be modified by the dicarbonyl group of AMPS-HDB. Arg174 is a strictly conserved residue among FDHs and is located at the Rossmann fold, the common mononucleotide-binding motif of dehydrogenases. Arg174 was replaced by Asn, using site-directed mutagenesis. The mutant enzyme CbFDHArg174Asn was showed to be resistant to inactivation by AMPS-HDB, confirming that the guanidinium group of Arg174 is the target for AMPS-HDB. The CbFDHArg174Asn mutant enzyme exhibited substantial reduced affinity for NAD+ and lower thermostability. The results of the study underline the pivotal and multifunctional role of Arg174 in catalysis, coenzyme binding and structural stability of CbFDH.
Keywords: NAD+ binding site; formate dehydrogenase; site-directed mutagenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

