Amplicon-Based Next Generation Sequencing for Rapid Identification of Rickettsia and Ectoparasite Species from Entomological Surveillance in Thailand
- PMID: 33669463
- PMCID: PMC7920428
- DOI: 10.3390/pathogens10020215
Amplicon-Based Next Generation Sequencing for Rapid Identification of Rickettsia and Ectoparasite Species from Entomological Surveillance in Thailand
Abstract
Background: Next generation sequencing (NGS) technology has been used for a wide range of epidemiological and surveillance studies. Here, we used amplicon-based NGS to species identify Rickettsia and their arthropod hosts from entomological surveillance.
Methods: During 2015-2016, we screened 1825 samples of rodents and ectoparasites collected from rodents and domestic mammals (dog, cat, and cattle) across Thailand for Rickettsia. The citrate synthase gene was amplified to identify Rickettsia to species, while the Cytochrome Oxidase subunit I (COI) and subunit II (COII) genes were used as target genes for ectoparasite identification. All target gene amplicons were pooled for library preparation and sequenced with Illumina MiSeq platform.
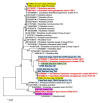
Result: The highest percentage of Rickettsia DNA was observed in fleas collected from domestic animals (56%) predominantly dogs. Only a few samples of ticks from domestic animals, rodent fleas, and rodent tissue were positive for Rickettisia DNA. NGS based characterization of Rickettsia by host identified Rickettsia asembonensis as the most common bacteria in positive fleas collected from dogs (83.2%) while "Candidatus Rickettsia senegalensis" was detected in only 16.8% of Rickettsia positive dog fleas. Sequence analysis of COI and COII revealed that almost all fleas collected from dogs were Ctenocephalides felis orientis. Other Rickettsia species were detected by NGS including Rickettsia heilongjiangensis from two Haemaphysalis hystricis ticks, and Rickettsia typhi in two rodent tissue samples.
Conclusion: This study demonstrates the utility of NGS for high-throughput sequencing in the species characterization/identification of bacteria and ectoparasite for entomological surveillance of rickettsiae. A high percentage of C. f. orientis are positive for R. asembonensis. In addition, our findings indicate there is a risk of tick-borne Spotted Fever Group rickettsiosis, and flea-borne murine typhus transmission in Tak and Phangnga provinces of Thailand.
Keywords: Ctenocephalides felis orientis; Next-generation sequencing (NGS); Rickettsia asembonensis.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

