Extensive Placental Methylation Profiling in Normal Pregnancies
- PMID: 33669975
- PMCID: PMC7924820
- DOI: 10.3390/ijms22042136
Extensive Placental Methylation Profiling in Normal Pregnancies
Erratum in
-
Correction: Rondinone et al. Extensive Placental Methylation Profiling in Normal Pregnancies. Int. J. Mol. Sci. 2021, 22, 2136.Int J Mol Sci. 2022 May 10;23(10):5298. doi: 10.3390/ijms23105298. Int J Mol Sci. 2022. PMID: 35628670 Free PMC article.
Abstract
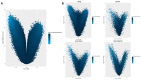
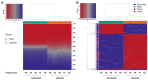
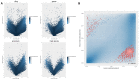
The placental methylation pattern is crucial for the regulation of genes involved in trophoblast invasion and placental development, both key events for fetal growth. We investigated LINE-1 methylation and methylome profiling using a methylation EPIC array and the targeted methylation sequencing of 154 normal, full-term pregnancies, stratified by birth weight percentiles. LINE-1 methylation showed evidence of a more pronounced hypomethylation in small neonates compared with normal and large for gestational age. Genome-wide methylation, performed in two subsets of pregnancies, showed very similar methylation profiles among cord blood samples while placentae from different pregnancies appeared very variable. A unique methylation profile emerged in each placenta, which could represent the sum of adjustments that the placenta made during the pregnancy to preserve the epigenetic homeostasis of the fetus. Investigations into the 1000 most variable sites between cord blood and the placenta showed that promoters and gene bodies that are hypermethylated in the placenta are associated with blood-specific functions, whereas those that are hypomethylated belong mainly to pathways involved in cancer. These features support the functional analogies between a placenta and cancer. Our results, which provide a comprehensive analysis of DNA methylation profiling in the human placenta, suggest that its peculiar dynamicity can be relevant for understanding placental plasticity in response to the environment.
Keywords: LINE-1; birth weight; methylome; normal pregnancies; placenta.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Miozzo M., Grati F., Bulfamante G., Rossella F., Cribiù M., Radaelli T., Cassani B., Persico T., Cetin I., Pardi G., et al. Post-zygotic origin of complete maternal chromosome 7 Isodisomy and consequent loss of placental PEG1/MEST expression. Placenta. 2001;22:813–821. doi: 10.1053/plac.2001.0728. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

