Exploring the Cold-Adaptation Mechanism of Serine Hydroxymethyltransferase by Comparative Molecular Dynamics Simulations
- PMID: 33670090
- PMCID: PMC7916883
- DOI: 10.3390/ijms22041781
Exploring the Cold-Adaptation Mechanism of Serine Hydroxymethyltransferase by Comparative Molecular Dynamics Simulations
Abstract
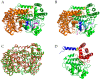
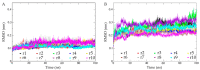
Cold-adapted enzymes feature a lower thermostability and higher catalytic activity compared to their warm-active homologues, which are considered as a consequence of increased flexibility of their molecular structures. The complexity of the (thermo)stability-flexibility-activity relationship makes it difficult to define the strategies and formulate a general theory for enzyme cold adaptation. Here, the psychrophilic serine hydroxymethyltransferase (pSHMT) from Psychromonas ingrahamii and its mesophilic counterpart, mSHMT from Escherichia coli, were subjected to μs-scale multiple-replica molecular dynamics (MD) simulations to explore the cold-adaptation mechanism of the dimeric SHMT. The comparative analyses of MD trajectories reveal that pSHMT exhibits larger structural fluctuations and inter-monomer positional movements, a higher global flexibility, and considerably enhanced local flexibility involving the surface loops and active sites. The largest-amplitude motion mode of pSHMT describes the trends of inter-monomer dissociation and enlargement of the active-site cavity, whereas that of mSHMT characterizes the opposite trends. Based on the comparison of the calculated structural parameters and constructed free energy landscapes (FELs) between the two enzymes, we discuss in-depth the physicochemical principles underlying the stability-flexibility-activity relationships and conclude that (i) pSHMT adopts the global-flexibility mechanism to adapt to the cold environment and, (ii) optimizing the protein-solvent interactions and loosening the inter-monomer association are the main strategies for pSHMT to enhance its flexibility.
Keywords: cold adaptation; free energy landscape; molecular dynamics simulation; protein-solvent interactions; stability-flexibility-activity relationships.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

