Identification and Validation of New Cancer Stem Cell-Related Genes and Their Regulatory microRNAs in Colorectal Cancerogenesis
- PMID: 33670246
- PMCID: PMC7916981
- DOI: 10.3390/biomedicines9020179
Identification and Validation of New Cancer Stem Cell-Related Genes and Their Regulatory microRNAs in Colorectal Cancerogenesis
Abstract
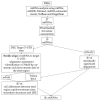
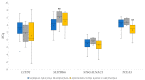
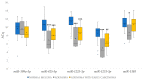
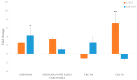
Significant progress has been made in the last decade in our understanding of the pathogenetic mechanisms of colorectal cancer (CRC). Cancer stem cells (CSC) have gained much attention and are now believed to play a crucial role in the pathogenesis of various cancers, including CRC. In the current study, we validated gene expression of four genes related to CSC, L1TD1, SLITRK6, ST6GALNAC1 and TCEA3, identified in a previous bioinformatics analysis. Using bioinformatics, potential miRNA-target gene correlations were prioritized. In total, 70 formalin-fixed paraffin-embedded biopsy samples from 47 patients with adenoma, adenoma with early carcinoma and CRC without and with lymph node metastases were included. The expression of selected genes and microRNAs (miRNAs) was evaluated using quantitative PCR. Differential expression of all investigated genes and four of six prioritized miRNAs (hsa-miR-199a-3p, hsa-miR-335-5p, hsa-miR-425-5p, hsa-miR-1225-3p, hsa-miR-1233-3p and hsa-miR-1303) was found in at least one group of CRC cancerogenesis. L1TD1, SLITRK6, miR-1233-3p and miR-1225-3p were correlated to the level of malignancy. A negative correlation between miR-199a-3p and its predicted target SLITRK6 was observed, showing potential for further experimental validation in CRC. Our results provide further evidence that CSC-related genes and their regulatory miRNAs are involved in CRC development and progression and suggest that some them, particularly miR-199a-3p and its SLITRK6 target gene, are promising for further validation in CRC.
Keywords: cancer stem cells; colorectal cancer; differentially expressed genes; qPCR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Intra‑tumor heterogeneity of cancer stem cell‑related genes and their potential regulatory microRNAs in metastasizing colorectal carcinoma.Oncol Rep. 2022 Nov;48(5):193. doi: 10.3892/or.2022.8408. Epub 2022 Sep 16. Oncol Rep. 2022. PMID: 36111489
-
MTUS1 and its targeting miRNAs in colorectal carcinoma: significant associations.Tumour Biol. 2016 May;37(5):6637-45. doi: 10.1007/s13277-015-4550-4. Epub 2015 Dec 7. Tumour Biol. 2016. PMID: 26643896
-
Identification of the differential expression of genes and upstream microRNAs in small cell lung cancer compared with normal lung based on bioinformatics analysis.Medicine (Baltimore). 2020 Mar;99(11):e19086. doi: 10.1097/MD.0000000000019086. Medicine (Baltimore). 2020. PMID: 32176034 Free PMC article.
-
Global and targeted circulating microRNA profiling of colorectal adenoma and colorectal cancer.Cancer. 2018 Feb 15;124(4):785-796. doi: 10.1002/cncr.31062. Epub 2017 Nov 7. Cancer. 2018. PMID: 29112225 Free PMC article.
-
MicroRNA dysregulation as a prognostic biomarker in colorectal cancer.Cancer Manag Res. 2014 Oct 14;6:405-22. doi: 10.2147/CMAR.S35164. eCollection 2014. Cancer Manag Res. 2014. PMID: 25342918 Free PMC article. Review.
Cited by
-
Bioinformatics Analysis of RNA-seq Data Reveals Genes Related to Cancer Stem Cells in Colorectal Cancerogenesis.Int J Mol Sci. 2022 Oct 31;23(21):13252. doi: 10.3390/ijms232113252. Int J Mol Sci. 2022. PMID: 36362041 Free PMC article.
-
The domesticated transposon protein L1TD1 associates with its ancestor L1 ORF1p to promote LINE-1 retrotransposition.Elife. 2025 Mar 20;13:RP96850. doi: 10.7554/eLife.96850. Elife. 2025. PMID: 40112032 Free PMC article.
-
Tumor-derived exosomal miR-425-5p and miR-135b-3p enhance colorectal cancer progression through immune suppression and vascular permeability promotion.World J Gastrointest Oncol. 2025 Jun 15;17(6):106161. doi: 10.4251/wjgo.v17.i6.106161. World J Gastrointest Oncol. 2025. PMID: 40547161 Free PMC article.
-
Development of a necroptosis-related prognostic model for uterine corpus endometrial carcinoma.Sci Rep. 2024 Feb 21;14(1):4257. doi: 10.1038/s41598-024-54651-3. Sci Rep. 2024. PMID: 38383747 Free PMC article.
-
Potential miRNA-gene interactions determining progression of various ATLL cancer subtypes after infection by HTLV-1 oncovirus.BMC Med Genomics. 2023 Mar 28;16(1):62. doi: 10.1186/s12920-023-01492-0. BMC Med Genomics. 2023. PMID: 36978083 Free PMC article.
References
-
- Kudryavtseva A.V., Lipatova A.V., Zaretsky A.R., Moskalev A.A., Fedorova M.S., Rasskazova A.S., Shibukhova G.A., Snezhkina A.V., Kaprin A.D., Alekseev B.Y., et al. Important molecular genetic markers of colorectal cancer. Oncotarget. 2016;7:53959–53983. doi: 10.18632/oncotarget.9796. - DOI - PMC - PubMed
-
- Sepulveda A.R., Portillo A.J.D. Molecular basis of diseases of the gastrointestinal tract. In: Coleman W.B., Tsongalis G.J., editors. Molecular Pathology. 2nd ed. Elsevier BV; Amsterdam, The Netherlands: 2018. pp. 387–415. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

