Berberine and Obatoclax Inhibit SARS-Cov-2 Replication in Primary Human Nasal Epithelial Cells In Vitro
- PMID: 33670363
- PMCID: PMC7918080
- DOI: 10.3390/v13020282
Berberine and Obatoclax Inhibit SARS-Cov-2 Replication in Primary Human Nasal Epithelial Cells In Vitro
Abstract
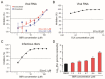
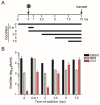
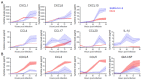
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged as a new human pathogen in late 2019 and it has infected over 100 million people in less than a year. There is a clear need for effective antiviral drugs to complement current preventive measures, including vaccines. In this study, we demonstrate that berberine and obatoclax, two broad-spectrum antiviral compounds, are effective against multiple isolates of SARS-CoV-2. Berberine, a plant-derived alkaloid, inhibited SARS-CoV-2 at low micromolar concentrations and obatoclax, which was originally developed as an anti-apoptotic protein antagonist, was effective at sub-micromolar concentrations. Time-of-addition studies indicated that berberine acts on the late stage of the viral life cycle. In agreement, berberine mildly affected viral RNA synthesis, but it strongly reduced infectious viral titers, leading to an increase in the particle-to-pfu ratio. In contrast, obatoclax acted at the early stage of the infection, which is in line with its activity to neutralize the acidic environment in endosomes. We assessed infection of primary human nasal epithelial cells that were cultured on an air-liquid interface and found that SARS-CoV-2 infection induced and repressed expression of specific sets of cytokines and chemokines. Moreover, both obatoclax and berberine inhibited SARS-CoV-2 replication in these primary target cells. We propose berberine and obatoclax as potential antiviral drugs against SARS-CoV-2 that could be considered for further efficacy testing.
Keywords: COVID-19; antiviral compounds; coronavirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

