Cold Adaptation in Antarctic Notothenioids: Comparative Transcriptomics Reveals Novel Insights in the Peculiar Role of Gills and Highlights Signatures of Cobalamin Deficiency
- PMID: 33670421
- PMCID: PMC7918649
- DOI: 10.3390/ijms22041812
Cold Adaptation in Antarctic Notothenioids: Comparative Transcriptomics Reveals Novel Insights in the Peculiar Role of Gills and Highlights Signatures of Cobalamin Deficiency
Abstract
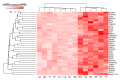
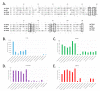
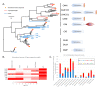
Far from being devoid of life, Antarctic waters are home to Cryonotothenioidea, which represent one of the fascinating cases of evolutionary adaptation to extreme environmental conditions in vertebrates. Thanks to a series of unique morphological and physiological peculiarities, which include the paradigmatic case of loss of hemoglobin in the family Channichthyidae, these fish survive and thrive at sub-zero temperatures. While some of the distinctive features of such adaptations have been known for decades, our knowledge of their genetic and molecular bases is still limited. We generated a reference de novo assembly of the icefish Chionodraco hamatus transcriptome and used this resource for a large-scale comparative analysis among five red-blooded Cryonotothenioidea, the sub-Antarctic notothenioid Eleginops maclovinus and seven temperate teleost species. Our investigation targeted the gills, a tissue of primary importance for gaseous exchange, osmoregulation, ammonia excretion, and its role in fish immunity. One hundred and twenty genes were identified as significantly up-regulated in Antarctic species and surprisingly shared by red- and white-blooded notothenioids, unveiling several previously unreported molecular players that might have contributed to the evolutionary success of Cryonotothenioidea in Antarctica. In particular, we detected cobalamin deficiency signatures and discussed the possible biological implications of this condition concerning hematological alterations and the heavy parasitic loads typically observed in all Cryonotothenioidea.
Keywords: Antarctica; Cryonotothenioidea; RNA-seq; cold adaptation; transcobalamin.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Cold-Driven Hemoglobin Evolution in Antarctic Notothenioid Fishes Prior to Hemoglobin Gene Loss in White-Blooded Icefishes.Mol Biol Evol. 2023 Nov 3;40(11):msad236. doi: 10.1093/molbev/msad236. Mol Biol Evol. 2023. PMID: 37879119 Free PMC article.
-
The evolutionary puzzle solution for the origins of the partial loss of the Cτ2 exon in notothenioid fishes.Fish Shellfish Immunol. 2021 Sep;116:124-139. doi: 10.1016/j.fsi.2021.05.015. Epub 2021 May 24. Fish Shellfish Immunol. 2021. PMID: 34038801
-
Biochemical adaptations of notothenioid fishes: comparisons between cold temperate South American and New Zealand species and Antarctic species.Comp Biochem Physiol A Mol Integr Physiol. 2007 Jul;147(3):799-807. doi: 10.1016/j.cbpa.2006.09.028. Epub 2006 Dec 5. Comp Biochem Physiol A Mol Integr Physiol. 2007. PMID: 17293146 Review.
-
Genome evolution in the cold: Antarctic icefish muscle transcriptome reveals selective duplications increasing mitochondrial function.Genome Biol Evol. 2013;5(1):45-60. doi: 10.1093/gbe/evs108. Genome Biol Evol. 2013. PMID: 23196969 Free PMC article.
-
Antarctic notothenioid fish: what are the future consequences of 'losses' and 'gains' acquired during long-term evolution at cold and stable temperatures?J Exp Biol. 2015 Jun;218(Pt 12):1834-45. doi: 10.1242/jeb.116129. J Exp Biol. 2015. PMID: 26085661 Review.
Cited by
-
Antarctic Fish as a Global Pollution Sensor: Metals Biomonitoring in a Twelve-Year Period.Front Mol Biosci. 2021 Dec 9;8:794946. doi: 10.3389/fmolb.2021.794946. eCollection 2021. Front Mol Biosci. 2021. PMID: 34957222 Free PMC article.
-
Gene loss in Antarctic icefish: evolutionary adaptations mimicking Fanconi Anemia?BMC Genomics. 2024 Nov 18;25(1):1102. doi: 10.1186/s12864-024-11028-0. BMC Genomics. 2024. PMID: 39558275 Free PMC article.
-
PTP-3 regulated by VB12 is important for ageing health in C. elegans.NPJ Aging. 2025 Feb 21;11(1):10. doi: 10.1038/s41514-025-00201-8. NPJ Aging. 2025. PMID: 39984496 Free PMC article.
References
-
- Hüne M., González-Wevar C., Poulin E., Mansilla A., Fernández D.A., Barrera-Oro E. Low Level of Genetic Divergence between Harpagifer Fish Species (Perciformes: Notothenioidei) Suggests a Quaternary Colonization of Patagonia from the Antarctic Peninsula. Polar Biol. 2015;38:607–617. doi: 10.1007/s00300-014-1623-6. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

