A Look into Bunyavirales Genomes: Functions of Non-Structural (NS) Proteins
- PMID: 33670641
- PMCID: PMC7922539
- DOI: 10.3390/v13020314
A Look into Bunyavirales Genomes: Functions of Non-Structural (NS) Proteins
Abstract
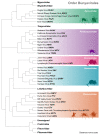
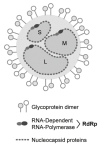
In 2016, the Bunyavirales order was established by the International Committee on Taxonomy of Viruses (ICTV) to incorporate the increasing number of related viruses across 13 viral families. While diverse, four of the families (Peribunyaviridae, Nairoviridae, Hantaviridae, and Phenuiviridae) contain known human pathogens and share a similar tri-segmented, negative-sense RNA genomic organization. In addition to the nucleoprotein and envelope glycoproteins encoded by the small and medium segments, respectively, many of the viruses in these families also encode for non-structural (NS) NSs and NSm proteins. The NSs of Phenuiviridae is the most extensively studied as a host interferon antagonist, functioning through a variety of mechanisms seen throughout the other three families. In addition, functions impacting cellular apoptosis, chromatin organization, and transcriptional activities, to name a few, are possessed by NSs across the families. Peribunyaviridae, Nairoviridae, and Phenuiviridae also encode an NSm, although less extensively studied than NSs, that has roles in antagonizing immune responses, promoting viral assembly and infectivity, and even maintenance of infection in host mosquito vectors. Overall, the similar and divergent roles of NS proteins of these human pathogenic Bunyavirales are of particular interest in understanding disease progression, viral pathogenesis, and developing strategies for interventions and treatments.
Keywords: bunyavirales; hantaviridae; interferon antagonist; nairoviridae; non-structural proteins; peribunyaviridae; phenuiviridae.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in data.
Figures

References
-
- Blitvich B.J., Beaty B.J., Blair C.D., Brault A.C., Dobler G., Drebot M.A., Haddow A.D., Kramer L.D., LaBeaud A.D., Monath T.P., et al. Bunyavirus Taxonomy: Limitations and Misconceptions Associated with the Current ICTV Criteria Used for Species Demarcation. Am. J. Trop. Med. Hyg. 2018;99:11–16. doi: 10.4269/ajtmh.18-0038. - DOI - PMC - PubMed
-
- CDC . Bunyaviridae. Centers for Disease Control and Prevention; Atlanta, GA, USA: 2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

