Cellular Fitness Phenotypes of Cancer Target Genes from Oncobiology to Cancer Therapeutics
- PMID: 33670680
- PMCID: PMC7921985
- DOI: 10.3390/cells10020433
Cellular Fitness Phenotypes of Cancer Target Genes from Oncobiology to Cancer Therapeutics
Abstract
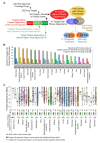
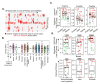
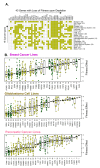
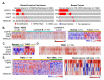
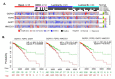
To define the growing significance of cellular targets and/or effectors of cancer drugs, we examined the fitness dependency of cellular targets and effectors of cancer drug targets across human cancer cells from 19 cancer types. We observed that the deletion of 35 out of 47 cellular effectors and/or targets of oncology drugs did not result in the expected loss of cell fitness in appropriate cancer types for which drugs targeting or utilizing these molecules for their actions were approved. Additionally, our analysis recognized 43 cellular molecules as fitness genes in several cancer types in which these drugs were not approved, and thus, providing clues for repurposing certain approved oncology drugs in such cancer types. For example, we found a widespread upregulation and fitness dependency of several components of the mevalonate and purine biosynthesis pathways (currently targeted by bisphosphonates, statins, and pemetrexed in certain cancers) and an association between the overexpression of these molecules and reduction in the overall survival duration of patients with breast and other hard-to-treat cancers, for which such drugs are not approved. In brief, the present analysis raised cautions about off-target and undesirable effects of certain oncology drugs in a subset of cancers where the intended cellular effectors of drug might not be good fitness genes and that this study offers a potential rationale for repurposing certain approved oncology drugs for targeted therapeutics in additional cancer types.
Keywords: Mevalonate and Purine biosynthesis; breast cancer hard-to-treat cancers; cancer fitness genes; oncology drugs; repurposing.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
New targets and targeted drugs for the treatment of cancer: an outlook to pediatric oncology.Pediatr Hematol Oncol. 2011 Oct;28(7):539-55. doi: 10.3109/08880018.2011.613094. Pediatr Hematol Oncol. 2011. PMID: 21936619 Review.
-
Repurposing non-oncology small-molecule drugs to improve cancer therapy: Current situation and future directions.Acta Pharm Sin B. 2022 Feb;12(2):532-557. doi: 10.1016/j.apsb.2021.09.006. Epub 2021 Sep 10. Acta Pharm Sin B. 2022. PMID: 35256933 Free PMC article. Review.
-
IMPACT web portal: oncology database integrating molecular profiles with actionable therapeutics.BMC Med Genomics. 2018 Apr 20;11(Suppl 2):26. doi: 10.1186/s12920-018-0350-1. BMC Med Genomics. 2018. PMID: 29697364 Free PMC article.
-
Illuminating biological pathways for drug targeting in head and neck squamous cell carcinoma.PLoS One. 2019 Oct 9;14(10):e0223639. doi: 10.1371/journal.pone.0223639. eCollection 2019. PLoS One. 2019. PMID: 31596908 Free PMC article.
-
Drug-repositioning opportunities for cancer therapy: novel molecular targets for known compounds.Drug Discov Today. 2016 Jan;21(1):190-199. doi: 10.1016/j.drudis.2015.09.017. Epub 2015 Oct 9. Drug Discov Today. 2016. PMID: 26456577 Review.
Cited by
-
Delineation of Pathogenomic Insights of Breast Cancer in Young Women.Cells. 2022 Jun 15;11(12):1927. doi: 10.3390/cells11121927. Cells. 2022. PMID: 35741056 Free PMC article.
-
Identification of the pyroptosis-related gene signature and risk score model for esophageal squamous cell carcinoma.Aging (Albany NY). 2023 Apr 17;15(8):3094-3106. doi: 10.18632/aging.204661. Epub 2023 Apr 17. Aging (Albany NY). 2023. PMID: 37071001 Free PMC article.
-
Prognostic Significance of Dysregulated Epigenomic and Chromatin Modifiers in Cervical Cancer.Cells. 2021 Oct 5;10(10):2665. doi: 10.3390/cells10102665. Cells. 2021. PMID: 34685645 Free PMC article.
-
Hyperactivation of p21-Activated Kinases in Human Cancer and Therapeutic Sensitivity.Biomedicines. 2023 Feb 5;11(2):462. doi: 10.3390/biomedicines11020462. Biomedicines. 2023. PMID: 36830998 Free PMC article. Review.
-
The Revelation of Continuously Organized, Co-Overexpressed Protein-Coding Genes with Roles in Cellular Communications in Breast Cancer.Cells. 2022 Nov 28;11(23):3806. doi: 10.3390/cells11233806. Cells. 2022. PMID: 36497066 Free PMC article.
References
-
- Lin A., Giuliano C.J., Palladino A., John K.M., Abramowicz C., Yuan M.L., Sausville E.L., Lukow D.A., Liu L., Chait A.R., et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 2019;11:eaaw8412. doi: 10.1126/scitranslmed.aaw8412. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

