Isolation of a New Infectious Pancreatic Necrosis Virus (IPNV) Variant from a Fish Farm in Scotland
- PMID: 33670941
- PMCID: PMC7997178
- DOI: 10.3390/v13030385
Isolation of a New Infectious Pancreatic Necrosis Virus (IPNV) Variant from a Fish Farm in Scotland
Abstract
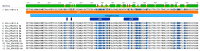
The aquatic virus, infectious pancreatic necrosis virus (IPNV), is known to infect various farmed fish, in particular salmonids, and is responsible for large economic losses in the aquaculture industry. Common practices to detect the virus include qPCR tests based on specific primers and serum neutralization tests for virus serotyping. Following the potential presence of IPNV viruses in a fish farm in Scotland containing vaccinated and IPNV-resistant fish, the common serotyping of the IPNV isolates was not made possible. This led us to determine the complete genome of the new IPNV isolates in order to investigate the cause of the serotyping discrepancy. Next-generation sequencing using the Illumina technology along with the sequence-independent single primer amplification (SISPA) approach was conducted to fully characterize the new Scottish isolates. With this approach, the full genome of two isolates, V1810-4 and V1810-6, was determined and analyzed. The potential origin of the virus isolates was investigated by phylogenetic analyses along with tridimensional and secondary protein structure analyses. These revealed the emergence of a new variant from one of the main virus serotypes, probably caused by the presence of selective pressure exerted by the vaccinated IPNV-resistant farmed fish.
Keywords: IPNV; aquaculture; evolution; infectious pancreatic necrosis virus; phylogeny; variant.
Conflict of interest statement
The authors declare no conflict of interests.
Figures



References
-
- Dobos P. The molecular biology of infectious pancreatic necrosis virus (IPNV) Annu. Rev. Fish Dis. 1995;5:25–54. doi: 10.1016/0959-8030(95)00003-8. - DOI
-
- Duncan R., Nagy E., Krell P.J., Dobos P.E. Synthesis of the infectious pancreatic necrosis virus polyprotein, detection of a virus-encoded protease, and fine structure mapping of genome segment A coding regions. J. Virol. 1987;61:3655–3664. doi: 10.1128/JVI.61.12.3655-3664.1987. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

