Immunogenomic Identification for Predicting the Prognosis of Cervical Cancer Patients
- PMID: 33671013
- PMCID: PMC7957482
- DOI: 10.3390/ijms22052442
Immunogenomic Identification for Predicting the Prognosis of Cervical Cancer Patients
Abstract
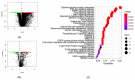
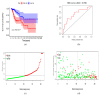
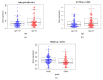
Cervical cancer is primarily caused by the infection of high-risk human papillomavirus (hrHPV). Moreover, tumor immune microenvironment plays a significant role in the tumorigenesis of cervical cancer. Therefore, it is necessary to comprehensively identify predictive biomarkers from immunogenomics associated with cervical cancer prognosis. The Cancer Genome Atlas (TCGA) public database has stored abundant sequencing or microarray data, and clinical data, offering a feasible and reliable approach for this study. In the present study, gene profile and clinical data were downloaded from TCGA, and the Immunology Database and Analysis Portal (ImmPort) database. Wilcoxon-test was used to compare the difference in gene expression. Univariate analysis was adopted to identify immune-related genes (IRGs) and transcription factors (TFs) correlated with survival. A prognostic prediction model was established by multivariate cox analysis. The regulatory network was constructed and visualized by correlation analysis and Cytoscape, respectively. Gene functional enrichment analysis was performed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). A total of 204 differentially expressed IRGs were identified, and 22 of them were significantly associated with the survival of cervical cancer. These 22 IRGs were actively involved in the JAK-STAT pathway. A prognostic model based on 10 IRGs (APOD, TFRC, GRN, CSK, HDAC1, NFATC4, BMP6, IL17RD, IL3RA, and LEPR) performed moderately and steadily in squamous cell carcinoma (SCC) patients with FIGO stage I, regardless of the age and grade. Taken together, a risk score model consisting of 10 novel genes capable of predicting survival in SCC patients was identified. Moreover, the regulatory network of IRGs associated with survival (SIRGs) and their TFs provided potential molecular targets.
Keywords: KEGG; TCGA; bioinformatics analysis; cervical cancer; tumor immune.
Conflict of interest statement
Sven Mahner reports grants and personal fees from AstraZeneca, personal fees from Clovis, grants, and personal fees from Medac, grants, and personal fees from MSD. He also reports personal fees from Novartis, grants and personal fees from PharmaMar, grants and personal fees from Roche, personal fees from Sensor Kinesis, grants, and personal fees from Tesaro, grants and personal fees from Teva, outside the submitted work. All other authors declare no conflict of interest.
Figures





References
-
- Ahn K., Kweon S., Kim D.W., Lee H. Different expression of GSK3beta and pS9GSK3beta depending on phenotype of cervical cancer: Possible association of GSK3beta with squamous cell carcinoma and pS9GSK3beta with adenocarcinoma. Obs. Gynecol. Sci. 2019;62:157–165. doi: 10.5468/ogs.2019.62.3.157. - DOI - PMC - PubMed
-
- van Meir H., Kenter G.G., Burggraaf J., Kroep J.R., Welters M.J., Melief C.J., Van Der Burg S.H., IE Van Poelgeest M. The need for improvement of the treatment of advanced and metastatic cervical cancer, the rationale for combined chemo-immunotherapy. Anticancer Agents Med. Chem. 2014;14:190–203. doi: 10.2174/18715206113136660372. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

