NGS-Based Application for Routine Non-Invasive Pre-Implantation Genetic Assessment in IVF
- PMID: 33671014
- PMCID: PMC7957524
- DOI: 10.3390/ijms22052443
NGS-Based Application for Routine Non-Invasive Pre-Implantation Genetic Assessment in IVF
Abstract
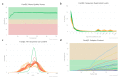
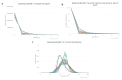
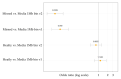
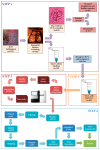
Although non-invasive pre-implantation genetic testing for aneuploidy (NIPGT-A) is potentially appropriate to assess chromosomal ploidy of the embryo, practical application of it in a routine IVF centre have not been started in the absence of a recommendation. Our objective in this study was to provide a comprehensive workflow for a clinically applicable strategy for NIPGT-A based on next-generation sequencing (NGS) technology with the corresponding bioinformatic pipeline. In a retrospective study, we performed NGS on spent blastocyst culture media of Day 3 embryos fertilised with intracytoplasmic sperm injection (ICSI) with quality score on morphology assessment using the blank culture media as background control. Chromosomal abnormalities were identified by an optimised bioinformatics pipeline applying copy number variation (CNV) detecting algorithm. In this study, we demonstrate a comprehensive workflow covering both wet- and dry-lab procedures supporting a clinically applicable strategy for NIPGT-A that can be carried out within 48 h, which is critical for the same-cycle blastocyst transfer. The described integrated approach of non-invasive evaluation of embryonic DNA content of the culture media can potentially supplement existing pre-implantation genetic screening methods.
Keywords: bioinformatic pipeline; in vitro fertilisation; missed abortion; next-generation sequencing; screening; spent culture medium.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
Relationship between age and blastocyst chromosomal ploidy analyzed by noninvasive preimplantation genetic testing for aneuploidies (niPGT-A).JBRA Assist Reprod. 2020 Oct 6;24(4):395-399. doi: 10.5935/1518-0557.20200061. JBRA Assist Reprod. 2020. PMID: 32723707 Free PMC article.
-
Minimizing mosaicism: assessing the impact of fertilization method on rate of mosaicism after next-generation sequencing (NGS) preimplantation genetic testing for aneuploidy (PGT-A).J Assist Reprod Genet. 2019 Jan;36(1):153-157. doi: 10.1007/s10815-018-1347-6. Epub 2018 Oct 25. J Assist Reprod Genet. 2019. PMID: 30362056 Free PMC article.
-
A pilot study to investigate the clinically predictive values of copy number variations detected by next-generation sequencing of cell-free deoxyribonucleic acid in spent culture media.Fertil Steril. 2024 Jul;122(1):42-51. doi: 10.1016/j.fertnstert.2024.02.030. Epub 2024 Feb 19. Fertil Steril. 2024. PMID: 38382698
-
Non-invasive preimplantation genetic testing (niPGT): the next revolution in reproductive genetics?Hum Reprod Update. 2020 Jan 1;26(1):16-42. doi: 10.1093/humupd/dmz033. Hum Reprod Update. 2020. PMID: 31774124 Review.
-
Non-Invasive Preimplantation Genetic Testing.Genes (Basel). 2025 Apr 30;16(5):552. doi: 10.3390/genes16050552. Genes (Basel). 2025. PMID: 40428374 Free PMC article. Review.
Cited by
-
Conditions for improved accuracy of noninvasive preimplantation genetic testing for aneuploidy: Focusing on the zona pellucida and early blastocysts.Reprod Med Biol. 2024 Sep 10;23(1):e12604. doi: 10.1002/rmb2.12604. eCollection 2024 Jan-Dec. Reprod Med Biol. 2024. PMID: 39263385 Free PMC article.
-
Epidemiological disease burden and annual, nationwide health insurance treatment cost of female infertility based on real-world health insurance claims data in Hungary.BMC Health Serv Res. 2025 Mar 4;25(1):336. doi: 10.1186/s12913-025-12348-x. BMC Health Serv Res. 2025. PMID: 40038705 Free PMC article.
-
Non-invasive preimplantation genetic testing for aneuploidy using cell-free DNA in blastocyst culture medium.J Assist Reprod Genet. 2025 May 21. doi: 10.1007/s10815-025-03510-9. Online ahead of print. J Assist Reprod Genet. 2025. PMID: 40399710
-
Non-Invasive Preimplantation Genetic Testing for Aneuploidy and the Mystery of Genetic Material: A Review Article.Int J Mol Sci. 2022 Mar 25;23(7):3568. doi: 10.3390/ijms23073568. Int J Mol Sci. 2022. PMID: 35408927 Free PMC article. Review.
-
Validation of Non-Invasive Preimplantation Genetic Screening Using a Routine IVF Laboratory Workflow.Biomedicines. 2022 Jun 11;10(6):1386. doi: 10.3390/biomedicines10061386. Biomedicines. 2022. PMID: 35740408 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

