High Risk Clone: A Proposal of Criteria Adapted to the One Health Context with Application to Enterotoxigenic Escherichia coli in the Pig Population
- PMID: 33671102
- PMCID: PMC8000703
- DOI: 10.3390/antibiotics10030244
High Risk Clone: A Proposal of Criteria Adapted to the One Health Context with Application to Enterotoxigenic Escherichia coli in the Pig Population
Abstract
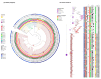
The definition of a high risk clone for antibiotic resistance dissemination was initially established for human medicine. We propose a revised definition of a high risk clone adapted to the One Health context. Then, we applied our criteria to a cluster of enrofloxacin non susceptible ETEC:F4 isolates which emerged in 2013 in diseased pigs in Quebec. The whole genomes of 183 ETEC:F4 strains isolated in Quebec from 1990 to 2018 were sequenced. The presence of virulence and resistance genes and replicons was examined in 173 isolates. Maximum likelihood phylogenetic trees were constructed based on SNP data and clones were identified using a set of predefined criteria. The strains belonging to the clonal lineage ST100/O149:H10 isolated in Quebec in 2013 or later were compared to ETEC:F4 whole genome sequences available in GenBank. Prior to 2000, ETEC:F4 isolates from pigs in Quebec were mostly ST90 and belonged to several serotypes. After 2000, the isolates were mostly ST100/O149:H10. In this article, we demonstrated the presence of a ETEC:F4 high risk clone. This clone (1) emerged in 2013, (2) is multidrug resistant, (3) has a widespread distribution over North America and was able to persist several months on farms, and (4) possesses specific virulence genes. It is crucial to detect and characterize high risk clones in animal populations to increase our understanding of their emergence and their dissemination.
Keywords: ETEC:F4; Escherichia coli; North America; antimicrobial resistance; fluoroquinolones non-susceptibility; genomics; multidrug resistance; pigs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
A new multidrug-resistant enterotoxigenic Escherichia coli pulsed-field gel electrophoresis cluster associated with enrofloxacin non-susceptibility in diseased pigs.J Appl Microbiol. 2021 Mar;130(3):707-721. doi: 10.1111/jam.14816. Epub 2020 Aug 25. J Appl Microbiol. 2021. PMID: 32767832 Free PMC article.
-
Isolation and characterization of nine bacteriophages that lyse O149 enterotoxigenic Escherichia coli.Vet Microbiol. 2007 Sep 20;124(1-2):47-57. doi: 10.1016/j.vetmic.2007.03.028. Epub 2007 Mar 30. Vet Microbiol. 2007. PMID: 17560053
-
Prevalence, Molecular Characterization, and Antimicrobial Resistance Profile of Enterotoxigenic Escherichia coli Isolates from Pig Farms in China.Foods. 2025 Mar 28;14(7):1188. doi: 10.3390/foods14071188. Foods. 2025. PMID: 40238372 Free PMC article.
-
Serotypes, virulence genes, and PFGE profiles of Escherichia coli isolated from pigs with postweaning diarrhoea in Slovakia.BMC Vet Res. 2006 Mar 20;2:10. doi: 10.1186/1746-6148-2-10. BMC Vet Res. 2006. PMID: 16549022 Free PMC article.
-
Methodology and application of Escherichia coli F4 and F18 encoding infection models in post-weaning pigs.J Anim Sci Biotechnol. 2019 Jun 13;10:53. doi: 10.1186/s40104-019-0352-7. eCollection 2019. J Anim Sci Biotechnol. 2019. PMID: 31210932 Free PMC article. Review.
Cited by
-
Description of Antimicrobial-Resistant Escherichia coli and Their Dissemination Mechanisms on Dairy Farms.Vet Sci. 2023 Mar 23;10(4):242. doi: 10.3390/vetsci10040242. Vet Sci. 2023. PMID: 37104397 Free PMC article.
-
Antimicrobial Resistance and Virulence Characteristics of Klebsiella pneumoniae Isolates in Kenya by Whole-Genome Sequencing.Pathogens. 2022 May 5;11(5):545. doi: 10.3390/pathogens11050545. Pathogens. 2022. PMID: 35631066 Free PMC article.
-
Genomic Profiling of Multidrug-Resistant Swine Escherichia coli and Clonal Relationship to Human Isolates in Peru.Antibiotics (Basel). 2023 Dec 18;12(12):1748. doi: 10.3390/antibiotics12121748. Antibiotics (Basel). 2023. PMID: 38136782 Free PMC article.
-
Impact of a Regulation Restricting Critical Antimicrobial Usage on Prevalence of Antimicrobial Resistance in Escherichia coli Isolates From Fecal and Manure Pit Samples on Dairy Farms in Québec, Canada.Front Vet Sci. 2022 Feb 17;9:838498. doi: 10.3389/fvets.2022.838498. eCollection 2022. Front Vet Sci. 2022. PMID: 35252426 Free PMC article.
-
Unveiling Antibiotic Resistance, Clonal Diversity, and Biofilm Formation in E. coli Isolated from Healthy Swine in Portugal.Pathogens. 2024 Apr 9;13(4):305. doi: 10.3390/pathogens13040305. Pathogens. 2024. PMID: 38668260 Free PMC article.
References
-
- Academies C.o.C. The Expert Panel on the Potential Socio-Economic Impacts of Antimicrobial Resistance in Canada. Council of Canadian Academies; Ottawa, ON, Canada: 2019. When antibiotics fail.
Grants and funding
- 37949/John-R. Evans Leaders Funds of the Canada foundation
- 116645/Ministère de l'agriculture, des pêcheries et de l'alimentation du Québec (MAPAQ), Programme Innov'Action
- RGPIN-2018-06560/Natural Sciences and Engineering Research Council of Canada (NSERC)
- 2016-047-C22/Consortium de recherche et innovations en bioprocédés industriels au Québec (CRIBIQ)
- CRIP Regroupements stratégiques 111946/Fonds de Recherche du Québec - Nature et Technologies
LinkOut - more resources
Full Text Sources
Other Literature Sources

