Mapping of Genomic Vulnerabilities in the Post-Translational Ubiquitination, SUMOylation and Neddylation Machinery in Breast Cancer
- PMID: 33671201
- PMCID: PMC7922122
- DOI: 10.3390/cancers13040833
Mapping of Genomic Vulnerabilities in the Post-Translational Ubiquitination, SUMOylation and Neddylation Machinery in Breast Cancer
Abstract
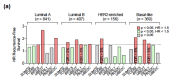
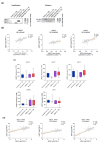
The dysregulation of post-translational modifications (PTM) transversally impacts cancer hallmarks and constitutes an appealing vulnerability for drug development. In breast cancer there is growing preclinical evidence of the role of ubiquitin and ubiquitin-like SUMO and Nedd8 peptide conjugation to the proteome in tumorigenesis and drug resistance, particularly through their interplay with estrogen receptor signaling and DNA repair. Herein we explored genomic alterations in these processes using RNA-seq and mutation data from TCGA and METABRIC datasets, and analyzed them using a bioinformatic pipeline in search of those with prognostic and predictive capability which could qualify as subjects of drug research. Amplification of UBE2T, UBE2C, and BIRC5 conferred a worse prognosis in luminal A/B and basal-like tumors, luminal A/B tumors, and luminal A tumors, respectively. Higher UBE2T expression levels were predictive of a lower rate of pathological complete response in triple negative breast cancer patients following neoadjuvant chemotherapy, whereas UBE2C and BIRC5 expression was higher in luminal A patients with tumor relapse within 5 years of endocrine therapy or chemotherapy. The transcriptomic signatures of USP9X and USP7 gene mutations also conferred worse prognosis in luminal A, HER2-enriched, and basal-like tumors, and in luminal A tumors, respectively. In conclusion, we identified and characterized the clinical value of a group of genomic alterations in ubiquitination, SUMOylation, and neddylation enzymes, with potential for drug development in breast cancer.
Keywords: SUMOylation; biomarkers; breast cancer; neddylation; post-translational modification; prognosis; ubiquitination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- Lord S.J., Kiely B.E., Pearson S.-A., Daniels B., O’Connell D.L., Beith J., Bulsara M.K., Houssami N. Metastatic Breast Cancer Incidence, Site and Survival in Australia, 2001–2016: A Population-Based Health Record Linkage Study Protocol. BMJ Open. 2019;9 doi: 10.1136/bmjopen-2018-026414. - DOI - PMC - PubMed
-
- Pereira B., Chin S.-F., Rueda O.M., Vollan H.-K.M., Provenzano E., Bardwell H.A., Pugh M., Jones L., Russell R., Sammut S.-J., et al. The Somatic Mutation Profiles of 2433 Breast Cancers Refine Their Genomic and Transcriptomic Landscapes. Nat. Commun. 2016;7:11479. doi: 10.1038/ncomms11479. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

