Enhanced Production of the Mical Redox Domain for Enzymology and F-actin Disassembly Assays
- PMID: 33671465
- PMCID: PMC7922515
- DOI: 10.3390/ijms22041991
Enhanced Production of the Mical Redox Domain for Enzymology and F-actin Disassembly Assays
Abstract
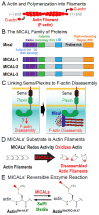
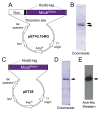
To change their behaviors, cells require actin proteins to assemble together into long polymers/filaments-and so a critical goal is to understand the factors that control this actin filament (F-actin) assembly and stability. We have identified a family of unusual actin regulators, the MICALs, which are flavoprotein monooxygenase/hydroxylase enzymes that associate with flavin adenine dinucleotide (FAD) and use the co-enzyme nicotinamide adenine dinucleotide phosphate (NADPH) in Redox reactions. F-actin is a specific substrate for these MICAL Redox enzymes, which oxidize specific amino acids within actin to destabilize actin filaments. Furthermore, this MICAL-catalyzed reaction is reversed by another family of Redox enzymes (SelR/MsrB enzymes)-thereby revealing a reversible Redox signaling process and biochemical mechanism regulating actin dynamics. Interestingly, in addition to the MICALs' Redox enzymatic portion through which MICALs covalently modify and affect actin, MICALs have multiple other domains. Less is known about the roles of these other MICAL domains. Here we provide approaches for obtaining high levels of recombinant protein for the Redox only portion of Mical and demonstrate its catalytic and F-actin disassembly activity. These results provide a ground state for future work aimed at defining the role of the other domains of Mical - including characterizing their effects on Mical's Redox enzymatic and F-actin disassembly activity.
Keywords: Drosophila; F-actin disassembly; MICAL1; MICAL2; MICAL3; oxidoreductase; plexin; repellent; repulsion; semaphorin.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
MICAL-mediated oxidation of actin and its effects on cytoskeletal and cellular dynamics.Front Cell Dev Biol. 2023 Feb 17;11:1124202. doi: 10.3389/fcell.2023.1124202. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36875759 Free PMC article. Review.
-
The MICALs are a Family of F-actin Dismantling Oxidoreductases Conserved from Drosophila to Humans.Sci Rep. 2018 Jan 17;8(1):937. doi: 10.1038/s41598-017-17943-5. Sci Rep. 2018. PMID: 29343822 Free PMC article.
-
Amplification of F-Actin Disassembly and Cellular Repulsion by Growth Factor Signaling.Dev Cell. 2017 Jul 24;42(2):117-129.e8. doi: 10.1016/j.devcel.2017.06.007. Epub 2017 Jul 6. Dev Cell. 2017. PMID: 28689759 Free PMC article.
-
Characterizing F-actin Disassembly Induced by the Semaphorin-Signaling Component MICAL.Methods Mol Biol. 2017;1493:119-128. doi: 10.1007/978-1-4939-6448-2_8. Methods Mol Biol. 2017. PMID: 27787846 Free PMC article.
-
Structure-function studies of MICAL, the unusual multidomain flavoenzyme involved in actin cytoskeleton dynamics.Arch Biochem Biophys. 2017 Oct 15;632:118-141. doi: 10.1016/j.abb.2017.06.004. Epub 2017 Jun 8. Arch Biochem Biophys. 2017. PMID: 28602956 Review.
Cited by
-
In Silico Identification of lncRNAs Regulating Sperm Motility in the Turkey (Meleagris gallopavo L.).Int J Mol Sci. 2022 Jul 11;23(14):7642. doi: 10.3390/ijms23147642. Int J Mol Sci. 2022. PMID: 35887003 Free PMC article.
-
MICAL-mediated oxidation of actin and its effects on cytoskeletal and cellular dynamics.Front Cell Dev Biol. 2023 Feb 17;11:1124202. doi: 10.3389/fcell.2023.1124202. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36875759 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

