Comparative Genomics: Insights on the Pathogenicity and Lifestyle of Rhizoctonia solani
- PMID: 33671736
- PMCID: PMC7926851
- DOI: 10.3390/ijms22042183
Comparative Genomics: Insights on the Pathogenicity and Lifestyle of Rhizoctonia solani
Abstract
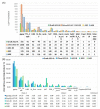
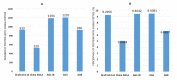
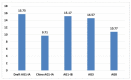
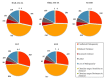
Proper management of agricultural disease is important to ensure sustainable food security. Staple food crops like rice, wheat, cereals, and other cash crops hold great export value for countries. Ensuring proper supply is critical; hence any biotic or abiotic factors contributing to the shortfall in yield of these crops should be alleviated. Rhizoctonia solani is a major biotic factor that results in yield losses in many agriculturally important crops. This paper focuses on genome informatics of our Malaysian Draft R. solani AG1-IA, and the comparative genomics (inter- and intra- AG) with four AGs including China AG1-IA (AG1-IA_KB317705.1), AG1-IB, AG3, and AG8. The genomic content of repeat elements, transposable elements (TEs), syntenic genomic blocks, functions of protein-coding genes as well as core orthologous genic information that underlies R. solani's pathogenicity strategy were investigated. Our analyses show that all studied AGs have low content and varying profiles of TEs. All AGs were dominant for Class I TE, much like other basidiomycete pathogens. All AGs demonstrate dominance in Glycoside Hydrolase protein-coding gene assignments suggesting its importance in infiltration and infection of host. Our profiling also provides a basis for further investigation on lack of correlation observed between number of pathogenicity and enzyme-related genes with host range. Despite being grouped within the same AG with China AG1-IA, our Draft AG1-IA exhibits differences in terms of protein-coding gene proportions and classifications. This implies that strains from similar AG do not necessarily have to retain similar proportions and classification of TE but must have the necessary arsenal to enable successful infiltration and colonization of host. In a larger perspective, all the studied AGs essentially share core genes that are generally involved in adhesion, penetration, and host colonization. However, the different infiltration strategies will depend on the level of host resilience where this is clearly exhibited by the gene sets encoded for the process of infiltration, infection, and protection from host.
Keywords: CAZy; Genome Annotation; Rhizoctonia solani; anastomosis group (AG); comparative genomics; pathogenicity genes; repeat element; single copy orthologs; synteny; transposable element.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Nagaraj B., Sunkad G., Pramesh D., Naik M., Patil M. Host range studies of rice sheath blight fungus Rhizoctonia solani (Kuhn) Int. J. Curr. Microbiol. App. Sci. 2017;6:3856–3864. doi: 10.20546/ijcmas.2017.611.452. - DOI
-
- García V.G., Onco M.P., Susan V.R. Biology and systematics of the form genus Rhizoctonia. Span. J. Agric. Res. 2006;4:55–79. doi: 10.5424/sjar/2006041-178. - DOI
-
- Wibberg D., Jelonek L., Rupp O., Hennig M., Eikmeyer F., Goesmann A., Hartmann A., Borriss R., Grosch R., Pühler A. Establishment and interpretation of the genome sequence of the phytopathogenic fungus Rhizoctonia solani AG1-IB isolate 7/3/14. J. Biotechnol. 2013;167:142–155. doi: 10.1016/j.jbiotec.2012.12.010. - DOI - PubMed
-
- Loan L., Du P., Li Z. Molecular dissection of quantitative resistance of sheath blight in rice (Oryza sativa L.) Omonrice. 2004;12:1–12.
-
- Pinson S.R., Capdevielle F.M., Oard J.H. Confirming QTLs and finding additional loci conditioning sheath blight resistance in rice using recombinant inbred lines. Crop Sci. 2005;45:503–510. doi: 10.2135/cropsci2005.0503. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

