Integrative Taxonomy and Molecular Phylogeny of the Plant-Parasitic Nematode Genus Paratylenchus (Nematoda: Paratylenchinae): Linking Species with Molecular Barcodes
- PMID: 33671787
- PMCID: PMC7926417
- DOI: 10.3390/plants10020408
Integrative Taxonomy and Molecular Phylogeny of the Plant-Parasitic Nematode Genus Paratylenchus (Nematoda: Paratylenchinae): Linking Species with Molecular Barcodes
Abstract
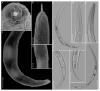
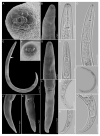
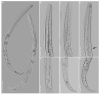
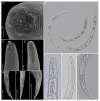
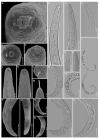
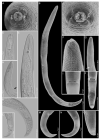
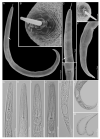
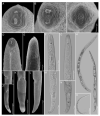
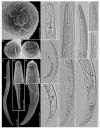
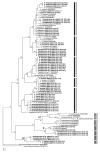
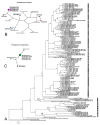
Pin nematodes of the genus Paratylenchus are obligate ectoparasites of a wide variety of plants that are distributed worldwide. In this study, individual morphologically vouchered nematode specimens of fourteen Paratylenchus species, including P. aculentus, P. elachistus, P. goodeyi, P. holdemani, P. idalimus, P. microdorus, P. nanus, P. neoamblycephalus, P. straeleni and P. veruculatus, are unequivocally linked to the D2-D3 of 28S, ITS, 18S rRNA and COI gene sequences. Combined with scanning electron microscopy and a molecular analysis of an additional nine known and thirteen unknown species originating from diverse geographic regions, a total of 92 D2-D3 of 28S, 41 ITS, 57 18S rRNA and 111 COI new gene sequences are presented. Paratylenchus elachistus, P. holdemani and P. neoamblycephalus are recorded for the first time in Belgium and P. idalimus for the first time in Europe. Paratylenchus is an excellent example of an incredibly diverse yet morphologically minimalistic plant-parasitic genus, and this study provides an integrated analysis of all available data, including coalescence-based molecular species delimitation, resulting in an updated Paratylenchus phylogeny and the corrective reassignment of 18 D2-D3 of 28S, 3 ITS, 3 18S rRNA and 25 COI gene sequences that were previously unidentified or incorrectly classified.
Keywords: 18S; COI; D2-D3 of 28S; ITS; Paratylenchus; morphology; morphometrics; phylogeny; plant-parasitic nematodes; taxonomy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Micoletzky H. Die freilebenden Erd-Nematoden. Archiv für Naturgeschichte; Berlin, Germany: 1922.
-
- Siddiqi M.R. Tylenchida: Parasites of Plants and Insects. 2nd ed. CABI Publishing; Wallingford, UK: 2000.
-
- Manzanilla-López R.H., Marbán-Mendoza N. Practical plant nematology. BBA; Colegio de Postgraduados, Mexico: 2012.
-
- Fourie H., Spaull V.W., Jones R.K., Daneel M.S., De Waele D. Nematology in South Africa: A View from the 21st Century. Springer; Cham, Switzerland: 2017.
-
- Subbotin S.A., Chitambar J.J. Plant Parasitic Nematodes in Sustainable Agriculture of North America. Springer; Cham, Switzerland: 2018.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

