Structure-Based Identification of Natural Products as SARS-CoV-2 Mpro Antagonist from Echinacea angustifolia Using Computational Approaches
- PMID: 33672054
- PMCID: PMC7919488
- DOI: 10.3390/v13020305
Structure-Based Identification of Natural Products as SARS-CoV-2 Mpro Antagonist from Echinacea angustifolia Using Computational Approaches
Abstract
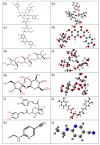
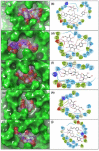
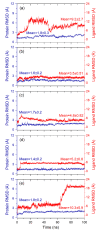
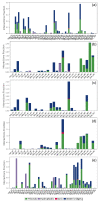
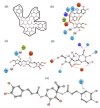
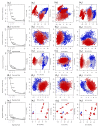
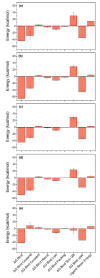
Coronavirus disease-19 (COVID-19) pandemic, caused by the novel SARS-CoV-2 virus, continues to be a global threat. The number of cases and deaths will remain escalating due to the lack of effective therapeutic agents. Several studies have established the importance of the viral main protease (Mpro) in the replication of SARS-CoV-2 which makes it an attractive target for antiviral drug development, including pharmaceutical repurposing and other medicinal chemistry approaches. Identification of natural products with considerable inhibitory potential against SARS-CoV-2 could be beneficial as a rapid and potent alternative with drug-likeness by comparison to de novo antiviral drug discovery approaches. Thereof, we carried out the structure-based screening of natural products from Echinacea-angustifolia, commonly used to prevent cold and other microbial respiratory infections, targeting SARS-CoV-2 Mpro. Four natural products namely, Echinacoside, Quercetagetin 7-glucoside, Levan N, Inulin from chicory, and 1,3-Dicaffeoylquinic acid, revealed significant docking energy (>-10 kcal/mol) in the SARS-CoV-2 Mpro catalytic pocket via substantial intermolecular contacts formation against co-crystallized ligand (<-4 kcal/mol). Furthermore, the docked poses of SARS-CoV-2 Mpro with selected natural products showed conformational stability through molecular dynamics. Exploring the end-point net binding energy exhibited substantial contribution of Coulomb and van der Waals interactions to the stability of respective docked conformations. These results advocated the natural products from Echinacea angustifolia for further experimental studies with an elevated probability to discover the potent SARS-CoV-2 Mpro antagonist with higher affinity and drug-likeness.
Keywords: COVID-19; Echinacea-angustifolia; Quercetagetin 7-glucoside; SARS-CoV-2; molecular dynamics simulation; natural products.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Targeting COVID-19 (SARS-CoV-2) main protease through active phytochemicals of ayurvedic medicinal plants - Withania somnifera (Ashwagandha), Tinospora cordifolia (Giloy) and Ocimum sanctum (Tulsi) - a molecular docking study.J Biomol Struct Dyn. 2022 Jan;40(1):190-203. doi: 10.1080/07391102.2020.1810778. Epub 2020 Aug 27. J Biomol Struct Dyn. 2022. PMID: 32851919 Free PMC article.
-
Computational Screening Using a Combination of Ligand-Based Machine Learning and Molecular Docking Methods for the Repurposing of Antivirals Targeting the SARS-CoV-2 Main Protease.Daru. 2024 Jun;32(1):47-65. doi: 10.1007/s40199-023-00484-w. Epub 2023 Oct 31. Daru. 2024. PMID: 37907683 Free PMC article.
-
An Updated Review on SARS-CoV-2 Main Proteinase (MPro): Protein Structure and Small-Molecule Inhibitors.Curr Top Med Chem. 2021;21(6):442-460. doi: 10.2174/1568026620666201207095117. Curr Top Med Chem. 2021. PMID: 33292134 Review.
-
Structure-based identification of SARS-CoV-2 main protease inhibitors from anti-viral specific chemical libraries: an exhaustive computational screening approach.Mol Divers. 2021 Aug;25(3):1979-1997. doi: 10.1007/s11030-021-10214-6. Epub 2021 Apr 12. Mol Divers. 2021. PMID: 33844135 Free PMC article.
-
SARS-CoV-2 Mpro: A Potential Target for Peptidomimetics and Small-Molecule Inhibitors.Biomolecules. 2021 Apr 19;11(4):607. doi: 10.3390/biom11040607. Biomolecules. 2021. PMID: 33921886 Free PMC article. Review.
Cited by
-
Computational exploration of the dual role of the phytochemical fortunellin: Antiviral activities against SARS-CoV-2 and immunomodulatory abilities against the host.Comput Biol Med. 2022 Oct;149:106049. doi: 10.1016/j.compbiomed.2022.106049. Epub 2022 Sep 8. Comput Biol Med. 2022. PMID: 36103744 Free PMC article.
-
Computational and In Vitro Experimental Investigations Reveal Anti-Viral Activity of Licorice and Glycyrrhizin against Severe Acute Respiratory Syndrome Coronavirus 2.Pharmaceuticals (Basel). 2021 Nov 24;14(12):1216. doi: 10.3390/ph14121216. Pharmaceuticals (Basel). 2021. PMID: 34959616 Free PMC article.
-
Alpha and gamma mangostins inhibit wild-type B SARS-CoV-2 more effectively than the SARS-CoV-2 variants and the major target is unlikely the 3C-like protease.Heliyon. 2024 May 27;10(11):e31987. doi: 10.1016/j.heliyon.2024.e31987. eCollection 2024 Jun 15. Heliyon. 2024. PMID: 38867992 Free PMC article.
-
Efficacy of Remdesivir and Neutralizing Monoclonal Antibodies in Monotherapy or Combination Therapy in Reducing the Risk of Disease Progression in Elderly or Immunocompromised Hosts Hospitalized for COVID-19: A Single Center Retrospective Study.Viruses. 2023 May 19;15(5):1199. doi: 10.3390/v15051199. Viruses. 2023. PMID: 37243285 Free PMC article.
-
Potent Bioactive Compounds From Seaweed Waste to Combat Cancer Through Bioinformatics Investigation.Front Nutr. 2022 Apr 22;9:889276. doi: 10.3389/fnut.2022.889276. eCollection 2022. Front Nutr. 2022. PMID: 35529456 Free PMC article.
References
-
- Coronavirus N. Novel Coronavirus (2019-nCoV) Situation Reports—World Health Organization. WHO; Geneva, Switzerland: 2020.
-
- Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., et al. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: Structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

