Investigation of Nasal/Oropharyngeal Microbial Community of COVID-19 Patients by 16S rDNA Sequencing
- PMID: 33672177
- PMCID: PMC7926517
- DOI: 10.3390/ijerph18042174
Investigation of Nasal/Oropharyngeal Microbial Community of COVID-19 Patients by 16S rDNA Sequencing
Abstract
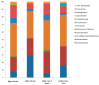
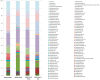
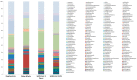
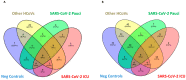
Since December 2019, SARS-CoV-2 infection has been still rapidly spreading, resulting in a pandemic, followed by an increasing number of cases in countries throughout the world. The severity of the disease depends on the patient's overall medical condition but no appropriate markers are available to establish the prognosis of the patients. We performed a 16S rRNA gene sequencing, revealing an altered composition of the nasal/oropharyngeal (NOP) microbiota in 21 patients affected by COVID-19, paucisymptomatic or in an Intensive Care Unit (ICU), as compared to 10 controls negative for COVID-19 or eight affected by a different Human Coronavirus (HKU, NL63 and OC43). A significant decrease in Chao1 index was observed when patients affected by COVID-19 (in ICU) were compared to paucisymptomatic. Furthermore, patients who were in ICU, paucisymptomatic or affected by other Coronaviruses all displayed a decrease in the Chao1 index when compared to controls, while Shannon index significantly decreased only in patients under ICU as compared to controls and paucisymptomatic patients. At the phylum level, Deinococcus-Thermus was present only in controls as compared to SARS-CoV-2 patients admitted to ICU, paucisymptomatic or affected by other coronaviruses. Candidatus Saccharibacteria (formerly known as TM7) was strongly increased in negative controls and SARS-CoV-2 paucisymptomatic patients as compared to SARS-CoV-2 ICU patients. Other modifications were observed at a lower taxonomy level. Complete depletion of Bifidobacterium and Clostridium was exclusively observed in ICU SARS-CoV-2 patients, which was the only group characterized by the presence of Salmonella, Scardovia, Serratia and Pectobacteriaceae. In conclusion, our preliminary results showed that nasal/oropharyngeal microbiota profiles of patients affected with SARS-CoV-2 may provide valuable information in order to facilitate the stratification of patients and may open the way to new interventional strategies in order to ameliorate the outcome of the patients.
Keywords: Nasal/Oropharyngeal; SARS-CoV2; microbiota.
Conflict of interest statement
All authors declare that they have no conflict of interest.
Figures

References
-
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Liu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. - DOI - PMC - PubMed
-
- Xu X.W., Wu X.X., Jiang X.G., Xu K.J., Ying L.J., Ma C.L., Li S.-B., Wang H.-Y., Zhang S., Gao H.-N., et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: Retrospective case series. BMJ. 2020;368:m606. doi: 10.1136/bmj.m606. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

