Concentration and Quantification of SARS-CoV-2 RNA in Wastewater Using Polyethylene Glycol-Based Concentration and qRT-PCR
- PMID: 33672247
- PMCID: PMC8005995
- DOI: 10.3390/mps4010017
Concentration and Quantification of SARS-CoV-2 RNA in Wastewater Using Polyethylene Glycol-Based Concentration and qRT-PCR
Erratum in
-
Correction: Farkas et al. Concentration and Quantification of SARS-CoV-2 RNA in Wastewater Using Polyethylene Glycol-Based Concentration and qRT-PCR. Methods Protoc. 2021, 4, 17.Methods Protoc. 2021 Nov 12;4(4):82. doi: 10.3390/mps4040082. Methods Protoc. 2021. PMID: 34842792 Free PMC article.
Abstract
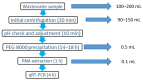
Wastewater-based epidemiology has become an important tool for the surveillance of SARS-CoV-2 outbreaks. However, the detection of viruses in sewage is challenging and to date there is no standard method available which has been validated for the sensitive detection of SARS-CoV-2. In this paper, we describe a simple concentration method based on polyethylene glycol (PEG) precipitation, followed by RNA extraction and a one-step quantitative reverse transcription PCR (qRT-PCR) for viral detection in wastewater. PEG-based concentration of viruses is a simple procedure which is not limited by the availability of expensive equipment and has reduced risk of disruption to consumable supply chains. The concentration and RNA extraction steps enable 900-1500× concentration of wastewater samples and sufficiently eliminates the majority of organic matter, which could inhibit the subsequent qRT-PCR assay. Due to the high variation in the physico-chemical properties of wastewater samples, we recommend the use of process control viruses to determine the efficiency of each step. This procedure enables the concentration and the extraction the DNA/RNA of different viruses and hence can be used for the surveillance of different viral targets for the comprehensive assessment of viral diseases in a community.
Keywords: COVID-19; environmental samples; public health; sewage; wastewater virology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Farkas K., Cooper D.M., McDonald J.E., Malham S.K., de Rougemont A., Jones D.L., de Rougemont A., Jones D.L., de Rougemont A., Jones D.L. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 2018;634:1174–1183. doi: 10.1016/j.scitotenv.2018.04.038. - DOI - PubMed
-
- Bisseux M., Colombet J., Mirand A., Roque-Afonso A.M., Abravanel F., Izopet J., Archimbaud C., Peigue-Lafeuille H., Debroas D., Bailly J.L., et al. Monitoring human enteric viruses in wastewater and relevance to infections encountered in the clinical setting: A one-year experiment in central France, 2014 to 2015. Eurosurveillance. 2018;23:1–11. doi: 10.2807/1560-7917.ES.2018.23.7.17-00237. - DOI - PMC - PubMed
-
- Ahmed W., Angel N., Edson J., Bibby K., Brien J.W.O., Choi P.M., Kitajima M., Simpson L., Li J., Tscharke B., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020:138764. doi: 10.1016/j.scitotenv.2020.138764. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous