IQGAP1 Is a Scaffold of the Core Proteins of the Hippo Pathway and Negatively Regulates the Pro-Apoptotic Signal Mediated by This Pathway
- PMID: 33672268
- PMCID: PMC7926663
- DOI: 10.3390/cells10020478
IQGAP1 Is a Scaffold of the Core Proteins of the Hippo Pathway and Negatively Regulates the Pro-Apoptotic Signal Mediated by This Pathway
Abstract
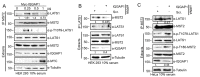
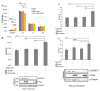
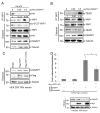
The Hippo pathway regulates a complex signalling network which mediates several biological functions including cell proliferation, organ size and apoptosis. Several scaffold proteins regulate the crosstalk of the members of the pathway with other signalling pathways and play an important role in the diverse output controlled by this pathway. In this study we have identified the scaffold protein IQGAP1 as a novel interactor of the core kinases of the Hippo pathway, MST2 and LATS1. Our results indicate that IQGAP1 scaffolds MST2 and LATS1 supresses their kinase activity and YAP1-dependent transcription. Additionally, we show that IQGAP1 is a negative regulator of the non-canonical pro-apoptotic pathway and may enable the crosstalk between this pathway and the ERK and AKT signalling modules. Our data also show that bile acids regulate the IQGAP1-MST2-LATS1 signalling module in hepatocellular carcinoma cells, which could be necessary for the inhibition of MST2-dependent apoptosis and hepatocyte transformation.
Keywords: Hippo; IQGAP1; LATS1; MST2; YAP1; bile acid; hepatocellular carcinoma.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Matallanas D., Romano D., Yee K., Meissl K., Kucerova L., Piazzolla D., Baccarini M., Vass J.K., Kolch W., O’Neill E. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol. Cell. 2007;27:962–975. doi: 10.1016/j.molcel.2007.08.008. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

