Runx3 Induces a Cell Shape Change and Suppresses Migration and Metastasis of Melanoma Cells by Altering a Transcriptional Profile
- PMID: 33672337
- PMCID: PMC7926509
- DOI: 10.3390/ijms22042219
Runx3 Induces a Cell Shape Change and Suppresses Migration and Metastasis of Melanoma Cells by Altering a Transcriptional Profile
Abstract
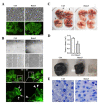
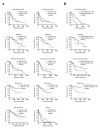
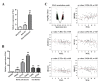
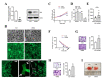
Runt-related transcription factor-3 (Runx3) is a tumor suppressor, and its contribution to melanoma progression remains unclear. We previously demonstrated that Runx3 re-expression in B16-F10 melanoma cells changed their shape and attenuated their migration. In this study, we found that Runx3 re-expression in B16-F10 cells also suppressed their pulmonary metastasis. We performed microarray analysis and uncovered an altered transcriptional profile underlying the cell shape change and the suppression of migration and metastasis. This altered transcriptional profile was rich in Gene Ontology/Kyoto Encyclopedia of Genes and Genomes (GO/KEGG) annotations relevant to adhesion and the actin cytoskeleton and included differentially expressed genes for some major extracellular matrix (ECM) proteins as well as genes that were inversely associated with the increase in the metastatic potential of B16-F10 cells compared to B16-F0 melanoma cells. Further, we found that this altered transcriptional profile could have prognostic value, as evidenced by myelin and lymphocyte protein (MAL) and vilin-like (VILL). Finally, Mal gene expression was correlated with metastatic potential among the cells and was targeted by histone deacetylase (HDAC) inhibitors in B16-F10 cells, and the knockdown of Mal gene expression in B16-F0 cells changed their shape and enhanced the migratory and invasive traits of their metastasis. Our study suggests that self-entrapping of metastatic Runx3-negative melanoma cells via adhesion and the actin cytoskeleton could be a powerful therapeutic strategy.
Keywords: Mal; Runx3; actin cytoskeleton; adhesion; extracellular matrix; melanoma; metastasis; transcriptional profile.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Runx3 inhibits melanoma cell migration through regulation of cell shape change.Cell Biol Int. 2017 Sep;41(9):1048-1055. doi: 10.1002/cbin.10824. Epub 2017 Jul 27. Cell Biol Int. 2017. PMID: 28699302
-
Protein 4.1B Suppresses Tumor Metastasis by Regulating Epithelial-mesenchymal Transition Progression in Melanoma Cells.Int J Med Sci. 2019 Apr 16;16(4):529-536. doi: 10.7150/ijms.27401. eCollection 2019. Int J Med Sci. 2019. PMID: 31171904 Free PMC article.
-
Identification of genes differentially expressed in B16 murine melanoma sublines with different metastatic potentials.Cancer Res. 1996 Feb 15;56(4):875-9. Cancer Res. 1996. PMID: 8631027
-
miR-301a promotes lung tumorigenesis by suppressing Runx3.Mol Cancer. 2019 May 23;18(1):99. doi: 10.1186/s12943-019-1024-0. Mol Cancer. 2019. PMID: 31122259 Free PMC article.
-
The emerging role of RUNX3 in cancer metastasis (Review).Oncol Rep. 2016 Mar;35(3):1227-36. doi: 10.3892/or.2015.4515. Epub 2015 Dec 24. Oncol Rep. 2016. PMID: 26708741 Review.
Cited by
-
RUNX transcription factors: biological functions and implications in cancer.Clin Exp Med. 2024 Mar 2;24(1):50. doi: 10.1007/s10238-023-01281-0. Clin Exp Med. 2024. PMID: 38430423 Free PMC article. Review.
-
Immune escape and metastasis mechanisms in melanoma: breaking down the dichotomy.Front Immunol. 2024 Feb 14;15:1336023. doi: 10.3389/fimmu.2024.1336023. eCollection 2024. Front Immunol. 2024. PMID: 38426087 Free PMC article. Review.
-
The role and mechanism of compressive stress in tumor.Front Oncol. 2024 Sep 16;14:1459313. doi: 10.3389/fonc.2024.1459313. eCollection 2024. Front Oncol. 2024. PMID: 39351360 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

