Understanding Rice- Magnaporthe Oryzae Interaction in Resistant and Susceptible Cultivars of Rice under Panicle Blast Infection Using a Time-Course Transcriptome Analysis
- PMID: 33672641
- PMCID: PMC7924189
- DOI: 10.3390/genes12020301
Understanding Rice- Magnaporthe Oryzae Interaction in Resistant and Susceptible Cultivars of Rice under Panicle Blast Infection Using a Time-Course Transcriptome Analysis
Abstract
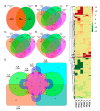
Rice blast is a global threat to food security with up to 50% yield losses. Panicle blast is a more severe form of rice blast and the response of rice plant to leaf and panicle blast is distinct in different genotypes. To understand the specific response of rice in panicle blast, transcriptome analysis of blast resistant cultivar Tetep, and susceptible cultivar HP2216 was carried out using RNA-Seq approach after 48, 72 and 96 h of infection with Magnaporthe oryzae along with mock inoculation. Transcriptome data analysis of infected panicle tissues revealed that 3553 genes differentially expressed in HP2216 and 2491 genes in Tetep, which must be the responsible factor behind the differential disease response. The defense responsive genes are involved mainly in defense pathways namely, hormonal regulation, synthesis of reactive oxygen species, secondary metabolites and cell wall modification. The common differentially expressed genes in both the cultivars were defense responsive transcription factors, NBS-LRR genes, kinases, pathogenesis related genes and peroxidases. In Tetep, cell wall strengthening pathway represented by PMR5, dirigent, tubulin, cell wall proteins, chitinases, and proteases was found to be specifically enriched. Additionally, many novel genes having DOMON, VWF, and PCaP1 domains which are specific to cell membrane were highly expressed only in Tetep post infection, suggesting their role in panicle blast resistance. Thus, our study shows that panicle blast resistance is a complex phenomenon contributed by early defense response through ROS production and detoxification, MAPK and LRR signaling, accumulation of antimicrobial compounds and secondary metabolites, and cell wall strengthening to prevent the entry and spread of the fungi. The present investigation provided valuable candidate genes that can unravel the mechanisms of panicle blast resistance and help in the rice blast breeding program.
Keywords: Magnaporthe; RNA-Seq; cell wall modification; disease resistance; panicle blast; rice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Sharma T.R., Rai A.K., Gupta S.K., Vijayan J., Devanna B.N., Ray S.K. Rice Blast Management Through Host-Plant Resistance: Retrospect and Prospects. Agric. Res. 2012;1:37–52. doi: 10.1007/s40003-011-0003-5. - DOI
-
- Asibi A.E., Chai Q., Coulter J.A. Rice Blast: A Disease with Implications for Global Food Security. Agronomy. 2019;9:451. doi: 10.3390/agronomy9080451. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

