Comprehensive Genome-Wide Exploration of C2H2 Zinc Finger Family in Grapevine (Vitis vinifera L.): Insights into the Roles in the Pollen Development Regulation
- PMID: 33672655
- PMCID: PMC7924211
- DOI: 10.3390/genes12020302
Comprehensive Genome-Wide Exploration of C2H2 Zinc Finger Family in Grapevine (Vitis vinifera L.): Insights into the Roles in the Pollen Development Regulation
Abstract
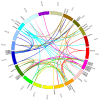
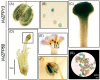
Some C2H2 zinc-finger proteins (ZFP) transcription factors are involved in the development of pollen in plants. In grapevine (Vitis vinifera L.), it has been suggested that abnormalities in pollen development lead to the phenomenon called parthenocarpy that occurs in some varieties of this cultivar. At present, a network involving several transcription factors types has been revealed and key roles have been assigned to members of the C2H2 zinc-finger proteins (ZFP) family in model plants. However, particularities of the regulatory mechanisms controlling pollen formation in grapevine remain unknown. In order to gain insight into the participation of ZFPs in grapevine gametophyte development, we performed a genome-wide identification and characterization of genes encoding ZFP (VviZFP family). A total of 98 genes were identified and renamed based on the gene distribution into grapevine genome. The analysis performed indicate significant changes throughout VviZFP genes evolution explained by high heterogeneity in sequence, length, number of ZF and presence of another conserved domains. Moreover, segmental duplication participated in the gene family expansion in grapevine. The VviZFPs were classified based on domain and phylogenetic analysis into three sets and different groups. Heat-map demonstrated differential and tissue-specific expression patterns of these genes and k-means clustering allowed to identify a group of putative orthologs to some ZFPs related to pollen development. In transgenic plants carrying the promVviZFP13::GUS and promVviZFP68::GUS constructs, GUS signals were detectable in the anther and mature pollen grains. Expression profiling of selected VviZFP genes showed differential expression pattern during flower development and provides a basis for deepening in the understanding of VviZFPs role on grapevine reproductive development.
Keywords: C2H2 zinc-finger protein; gene expression profiling; genome-wide; grapevine (Vitis vinifera L.); pollen development; transcription factor.
Conflict of interest statement
The authors declare that they have no conflicts of interest concerning this article.
Figures







Similar articles
-
Molecular characterization and expression analysis reveal the roles of Cys2/His2 zinc-finger transcription factors during flower development of Brassica rapa subsp. chinensis.Plant Mol Biol. 2020 Jan;102(1-2):123-141. doi: 10.1007/s11103-019-00935-6. Epub 2019 Nov 27. Plant Mol Biol. 2020. PMID: 31776846
-
Genome-Wide Identification and Analysis of the Genes Encoding Q-Type C2H2 Zinc Finger Proteins in Grapevine.Int J Mol Sci. 2023 Oct 14;24(20):15180. doi: 10.3390/ijms242015180. Int J Mol Sci. 2023. PMID: 37894862 Free PMC article.
-
Comprehensive genomic survey, structural classification and expression analysis of C2H2 zinc finger protein gene family in Brassica rapa L.PLoS One. 2019 May 6;14(5):e0216071. doi: 10.1371/journal.pone.0216071. eCollection 2019. PLoS One. 2019. PMID: 31059545 Free PMC article.
-
Genome-wide exploration of C2H2 zinc finger family in durum wheat (Triticum turgidum ssp. Durum): insights into the roles in biological processes especially stress response.Biometals. 2018 Dec;31(6):1019-1042. doi: 10.1007/s10534-018-0146-y. Epub 2018 Oct 4. Biometals. 2018. PMID: 30288657 Review.
-
Cys₂/His₂ Zinc-Finger Proteins in Transcriptional Regulation of Flower Development.Int J Mol Sci. 2018 Aug 31;19(9):2589. doi: 10.3390/ijms19092589. Int J Mol Sci. 2018. PMID: 30200325 Free PMC article. Review.
Cited by
-
Genome-Wide Analysis of Q-Type C2H2 ZFP Genes in Response to Biotic and Abiotic Stresses in Sugar Beet.Biology (Basel). 2023 Oct 4;12(10):1309. doi: 10.3390/biology12101309. Biology (Basel). 2023. PMID: 37887019 Free PMC article.
-
Functional characterization of four soybean C2H2 zinc-finger genes in Phytophthora resistance.Plant Signal Behav. 2025 Dec;20(1):2481185. doi: 10.1080/15592324.2025.2481185. Epub 2025 Mar 20. Plant Signal Behav. 2025. PMID: 40110654 Free PMC article.
-
Self S-RNase reduces the expression of two pollen-specific COBRA genes to inhibit pollen tube growth in pear.Mol Hortic. 2023 Dec 1;3(1):26. doi: 10.1186/s43897-023-00074-z. Mol Hortic. 2023. PMID: 38037174 Free PMC article.
-
Genome-wide identification of the C2H2-Zinc finger gene family and functional validation of CsZFP7 in citrus nucellar embryogenesis.Plant Reprod. 2023 Dec;36(4):287-300. doi: 10.1007/s00497-023-00470-x. Epub 2023 May 29. Plant Reprod. 2023. PMID: 37247027
-
Advances in the Regulation of Epidermal Cell Development by C2H2 Zinc Finger Proteins in Plants.Front Plant Sci. 2021 Sep 24;12:754512. doi: 10.3389/fpls.2021.754512. eCollection 2021. Front Plant Sci. 2021. PMID: 34630497 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

