Effects of Irregular Feeding on the Daily Fluctuations in mRNA Expression of the Neurosecretory Protein GL and Neurosecretory Protein GM Genes in the Mouse Hypothalamus
- PMID: 33672695
- PMCID: PMC7924315
- DOI: 10.3390/ijms22042109
Effects of Irregular Feeding on the Daily Fluctuations in mRNA Expression of the Neurosecretory Protein GL and Neurosecretory Protein GM Genes in the Mouse Hypothalamus
Abstract
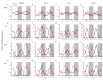
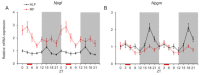
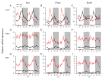
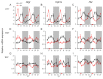
Circadian desynchrony induced by a long period of irregular feeding leads to metabolic diseases, such as obesity and diabetes mellitus. The recently identified neurosecretory protein GL (NPGL) and neurosecretory protein GM (NPGM) are hypothalamic small proteins that stimulate food intake and fat accumulation in several animals. To clarify the mechanisms that evoke feeding behavior and induce energy metabolism at the appropriate times in accordance with a circadian rhythm, diurnal fluctuations in Npgl and Npgm mRNA expression were investigated in mice. Quantitative RT-PCR analysis revealed that the mRNAs of these two genes were highly expressed in the mediobasal hypothalamus during the active dark phase under ad libitum feeding. In mice restricted to 3 h of feeding during the inactive light phase, the Npgl mRNA level was augmented in the moment prior to the feeding period and the midnight peak of Npgm mRNA was attenuated. Moreover, the mRNA expression levels of clock genes, feeding regulatory neuropeptides, and lipid metabolic enzymes in the central and peripheral tissues were comparable to those of central Npgl and Npgm. These data suggest that Npgl and Npgm transcription fluctuates daily and likely mediates feeding behavior and/or energy metabolism at an appropriate time according to the meal timing.
Keywords: environmental light/dark cycle; fat accumulation; feeding restriction; food intake; neurosecretory protein GL; neurosecretory protein GM; rhythmicity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Expression of mRNAs Encoding Hypothalamic Small Proteins, Neurosecretory Protein GL and Neurosecretory Protein GM, in the Japanese Quail, Coturnix japonica.Zoolog Sci. 2024 Feb;41(1):50-59. doi: 10.2108/zs230070. Zoolog Sci. 2024. PMID: 38587517
-
Localization and function of neurosecretory protein GM, a novel small secretory protein, in the chicken hypothalamus.Sci Rep. 2018 Jan 15;8(1):704. doi: 10.1038/s41598-017-18822-9. Sci Rep. 2018. PMID: 29335496 Free PMC article.
-
Hypothalamic Overexpression of Neurosecretory Protein GL Leads to Obesity in Male C57BL/6J Mice.Neuroendocrinology. 2022;112(6):606-620. doi: 10.1159/000518969. Epub 2021 Aug 12. Neuroendocrinology. 2022. PMID: 34384081
-
Avian and murine neurosecretory protein GL participates in the regulation of feeding and energy metabolism.Gen Comp Endocrinol. 2018 May 1;260:164-170. doi: 10.1016/j.ygcen.2017.09.019. Epub 2017 Sep 23. Gen Comp Endocrinol. 2018. PMID: 28951261 Review.
-
Circadian Rhythms in Diet-Induced Obesity.Adv Exp Med Biol. 2017;960:19-52. doi: 10.1007/978-3-319-48382-5_2. Adv Exp Med Biol. 2017. PMID: 28585194 Review.
Cited by
-
Sex-dependent circadian alterations of both central and peripheral clock genes expression and gut-microbiota composition during activity-based anorexia in mice.Biol Sex Differ. 2024 Jan 12;15(1):6. doi: 10.1186/s13293-023-00576-x. Biol Sex Differ. 2024. PMID: 38217033 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

