Targeting SHIP1 and SHIP2 in Cancer
- PMID: 33672717
- PMCID: PMC7924360
- DOI: 10.3390/cancers13040890
Targeting SHIP1 and SHIP2 in Cancer
Abstract
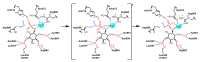
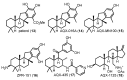
Membrane-anchored and soluble inositol phospholipid species are critical mediators of intracellular cell signaling cascades. Alterations in their normal production or degradation are implicated in the pathology of a number of disorders including cancer and pro-inflammatory conditions. The SH2-containing 5' inositol phosphatases, SHIP1 and SHIP2, play a fundamental role in these processes by depleting PI(3,4,5)P3, but also by producing PI(3,4)P2 at the inner leaflet of the plasma membrane. With the intent of targeting SHIP1 or SHIP2 selectively, or both paralogs simultaneously, small molecule inhibitors and agonists have been developed and tested in vitro and in vivo over the last decade in various disease models. These studies have shown promising results in various pre-clinical models of disease including cancer and tumor immunotherapy. In this review the potential use of SHIP inhibitors in cancer is discussed with particular attention to the molecular structure, binding site and efficacy of these SHIP inhibitors.
Keywords: AKT; Caspase 8; Fas; PI(3,4)P2; PI(3,4,5)P3; PI3K; SHIP1; SHIP2; SHIPi; cancer.
Conflict of interest statement
W.G.K., C.P. and J.D.C. have patents on small molecules targeting of SHIP1 and SHIP2 in disease. W.G.K. is Chief Scientific Officer and J.D.C. serves on the Scientific Advisory Board of the company “Alterna Therapeutics” for development and commercialization of SHIP inhibitor therapeutics. The other authors have no conflicts to disclose.
Figures


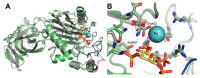
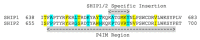


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

