Genomic Prediction Using Alternative Strategies of Weighted Single-Step Genomic BLUP for Yearling Weight and Carcass Traits in Hanwoo Beef Cattle
- PMID: 33673102
- PMCID: PMC7917987
- DOI: 10.3390/genes12020266
Genomic Prediction Using Alternative Strategies of Weighted Single-Step Genomic BLUP for Yearling Weight and Carcass Traits in Hanwoo Beef Cattle
Abstract
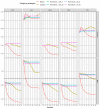
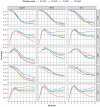
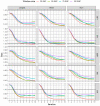
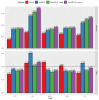
The weighted single-step genomic best linear unbiased prediction (GBLUP) method has been proposed to exploit information from genotyped and non-genotyped relatives, allowing the use of weights for single-nucleotide polymorphism in the construction of the genomic relationship matrix. The purpose of this study was to investigate the accuracy of genetic prediction using the following single-trait best linear unbiased prediction methods in Hanwoo beef cattle: pedigree-based (PBLUP), un-weighted (ssGBLUP), and weighted (WssGBLUP) single-step genomic methods. We also assessed the impact of alternative single and window weighting methods according to their effects on the traits of interest. The data was comprised of 15,796 phenotypic records for yearling weight (YW) and 5622 records for carcass traits (backfat thickness: BFT, carcass weight: CW, eye muscle area: EMA, and marbling score: MS). Also, the genotypic data included 6616 animals for YW and 5134 for carcass traits on the 43,950 single-nucleotide polymorphisms. The ssGBLUP showed significant improvement in genomic prediction accuracy for carcass traits (71%) and yearling weight (99%) compared to the pedigree-based method. The window weighting procedures performed better than single SNP weighting for CW (11%), EMA (11%), MS (3%), and YW (6%), whereas no gain in accuracy was observed for BFT. Besides, the improvement in accuracy between window WssGBLUP and the un-weighted method was low for BFT and MS, while for CW, EMA, and YW resulted in a gain of 22%, 15%, and 20%, respectively, which indicates the presence of relevant quantitative trait loci for these traits. These findings indicate that WssGBLUP is an appropriate method for traits with a large quantitative trait loci effect.
Keywords: Hanwoo cattle; SNP window; carcass traits; weighted single-step genomic procedures; yearling weight.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Multi-Trait Single-Step GBLUP Improves Accuracy of Genomic Prediction for Carcass Traits Using Yearling Weight and Ultrasound Traits in Hanwoo.Front Genet. 2021 Jul 30;12:692356. doi: 10.3389/fgene.2021.692356. eCollection 2021. Front Genet. 2021. PMID: 34394186 Free PMC article.
-
Exploring and Identifying Candidate Genes and Genomic Regions Related to Economically Important Traits in Hanwoo Cattle.Curr Issues Mol Biol. 2022 Dec 4;44(12):6075-6092. doi: 10.3390/cimb44120414. Curr Issues Mol Biol. 2022. PMID: 36547075 Free PMC article. Review.
-
Comparison of conventional BLUP and single-step genomic BLUP evaluations for yearling weight and carcass traits in Hanwoo beef cattle using single trait and multi-trait models.PLoS One. 2019 Oct 14;14(10):e0223352. doi: 10.1371/journal.pone.0223352. eCollection 2019. PLoS One. 2019. PMID: 31609979 Free PMC article.
-
Improving the accuracy of genomic evaluation for linear body measurement traits using single-step genomic best linear unbiased prediction in Hanwoo beef cattle.BMC Genet. 2020 Dec 2;21(1):144. doi: 10.1186/s12863-020-00928-1. BMC Genet. 2020. PMID: 33267771 Free PMC article.
-
Current status of genomic evaluation.J Anim Sci. 2020 Apr 1;98(4):skaa101. doi: 10.1093/jas/skaa101. J Anim Sci. 2020. PMID: 32267923 Free PMC article. Review.
Cited by
-
Haplotype-Based Single-Step GWAS for Yearling Temperament in American Angus Cattle.Genes (Basel). 2021 Dec 22;13(1):17. doi: 10.3390/genes13010017. Genes (Basel). 2021. PMID: 35052358 Free PMC article.
-
Multi-Trait Single-Step GBLUP Improves Accuracy of Genomic Prediction for Carcass Traits Using Yearling Weight and Ultrasound Traits in Hanwoo.Front Genet. 2021 Jul 30;12:692356. doi: 10.3389/fgene.2021.692356. eCollection 2021. Front Genet. 2021. PMID: 34394186 Free PMC article.
-
Exploring and Identifying Candidate Genes and Genomic Regions Related to Economically Important Traits in Hanwoo Cattle.Curr Issues Mol Biol. 2022 Dec 4;44(12):6075-6092. doi: 10.3390/cimb44120414. Curr Issues Mol Biol. 2022. PMID: 36547075 Free PMC article. Review.
-
Estimation of genetic correlations and genomic prediction accuracy for reproductive and carcass traits in Hanwoo cows.J Anim Sci Technol. 2024 Jul;66(4):682-701. doi: 10.5187/jast.2024.e75. Epub 2024 Jul 31. J Anim Sci Technol. 2024. PMID: 39165742 Free PMC article.
-
Pleiotropic Genes Affecting Milk Production, Fertility, and Health in Thai-Holstein Crossbred Dairy Cattle: A GWAS Approach.Animals (Basel). 2025 May 2;15(9):1320. doi: 10.3390/ani15091320. Animals (Basel). 2025. PMID: 40362136 Free PMC article.
References
-
- Wang H., Misztal I., Aguilar I., Legarra A., Fernando R.L., Vitezica Z., Okimoto R., Wing T., Hawken R., Muir W.M. Genome-wide association mapping including phenotypes from relatives without genotypes in a single-step (ssGWAS) for 6-week body weight in broiler chickens. Front. Genet. 2014;5:134. doi: 10.3389/fgene.2014.00134. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

