Analysis of the Conditions That Affect the Selective Processing of Endogenous Notch1 by ADAM10 and ADAM17
- PMID: 33673337
- PMCID: PMC7918056
- DOI: 10.3390/ijms22041846
Analysis of the Conditions That Affect the Selective Processing of Endogenous Notch1 by ADAM10 and ADAM17
Abstract
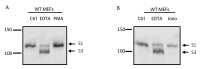
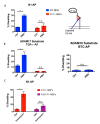
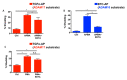
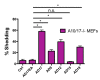
Notch signaling is critical for controlling a variety of cell fate decisions during metazoan development and homeostasis. This unique, highly conserved signaling pathway relies on cell-to-cell contact, which triggers the proteolytic release of the cytoplasmic domain of the membrane-anchored transcription factor Notch from the membrane. A disintegrin and metalloproteinase (ADAM) proteins are crucial for Notch activation by processing its S2 site. While ADAM10 cleaves Notch1 under physiological, ligand-dependent conditions, ADAM17 mainly cleaves Notch1 under ligand-independent conditions. However, the mechanism(s) that regulate the distinct contributions of these ADAMs in Notch processing remain unclear. Using cell-based assays in mouse embryonic fibroblasts (mEFs) lacking ADAM10 and/or ADAM17, we aimed to clarify what determines the relative contributions of ADAM10 and ADAM17 to ligand-dependent or ligand-independent Notch processing. We found that EDTA-stimulated ADAM17-dependent Notch1 processing is rapid and requires the ADAM17-regulators iRhom1 and iRhom2, whereas the Delta-like 4-induced ligand-dependent Notch1 processing is slower and requires ADAM10. The selectivity of ADAM17 for EDTA-induced Notch1 processing can most likely be explained by a preference for ADAM17 over ADAM10 for the Notch1 cleavage site and by the stronger inhibition of ADAM10 by EDTA. The physiological ADAM10-dependent processing of Notch1 cannot be compensated for by ADAM17 in Adam10-/- mEFs, or by other ADAMs shown here to be able to cleave the Notch1 cleavage site, such as ADAMs9, 12, and 19. Collectively, these results provide new insights into the mechanisms underlying the substrate selectivity of ADAM10 and ADAM17 towards Notch1.
Keywords: ADAM10; ADAM17; Notch pathway; Notch receptor; Notch1; cell signaling; intercellular signaling; juxtacrine signaling; proteolysis; regulation.
Conflict of interest statement
Carl Blobel holds a patent on a method of identifying agents for combination with inhibitors of iRhoms. Carl Blobel and the Hospital for Special Surgery have identified iRhom2 inhibitors and have co-founded the start-up company SciRhom in Munich to commercialize these inhibitors.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

