The Alter Retina: Alternative Splicing of Retinal Genes in Health and Disease
- PMID: 33673358
- PMCID: PMC7917623
- DOI: 10.3390/ijms22041855
The Alter Retina: Alternative Splicing of Retinal Genes in Health and Disease
Abstract
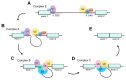
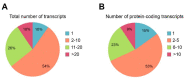
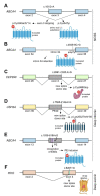
Alternative splicing of mRNA is an essential mechanism to regulate and increase the diversity of the transcriptome and proteome. Alternative splicing frequently occurs in a tissue- or time-specific manner, contributing to differential gene expression between cell types during development. Neural tissues present extremely complex splicing programs and display the highest number of alternative splicing events. As an extension of the central nervous system, the retina constitutes an excellent system to illustrate the high diversity of neural transcripts. The retina expresses retinal specific splicing factors and produces a large number of alternative transcripts, including exclusive tissue-specific exons, which require an exquisite regulation. In fact, a current challenge in the genetic diagnosis of inherited retinal diseases stems from the lack of information regarding alternative splicing of retinal genes, as a considerable percentage of mutations alter splicing or the relative production of alternative transcripts. Modulation of alternative splicing in the retina is also instrumental in the design of novel therapeutic approaches for retinal dystrophies, since it enables precision medicine for specific mutations.
Keywords: alternative splicing; deep intronic variants; inherited retinal dystrophies; microexons; non-canonical splice site variants; retina; splicing factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

