Dissecting the Gene Expression Networks Associated with Variations in the Major Components of the Fatty Acid Semimembranosus Muscle Profile in Large White Heavy Pigs
- PMID: 33673460
- PMCID: PMC7997476
- DOI: 10.3390/ani11030628
Dissecting the Gene Expression Networks Associated with Variations in the Major Components of the Fatty Acid Semimembranosus Muscle Profile in Large White Heavy Pigs
Abstract
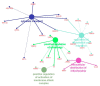
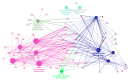
To date, high-throughput technology such as RNA-sequencing has been successfully applied in livestock sciences to investigate molecular networks involved in complex traits, such as meat quality. Pork quality depends on several organoleptic, technological, and nutritional characteristics, and it is also influenced by the fatty acid (FA) composition of intramuscular fat (IMF). To explore the molecular networks associated with different IMF FA compositions, the Semimembranosus muscle (SM) from two groups of Italian Large White (ILW) heavy pigs divergent for SM IMF content was investigated using transcriptome analysis. After alignment and normalization, the obtained gene counts were used to perform the Weighted Correlation Network Analysis (WGCNA package in R environment). Palmitic and palmitoleic contents showed association with the same gene modules, comprising genes significantly enriched in autophagy, mitochondrial fusion, and mitochondrial activity. Among the key genes related to these FAs, we found TEAD4, a gene regulating mitochondrial activity that seems to be a promising candidate for further studies. On the other hand, the genes comprised in the modules associated with the IMF contents of oleic, n-6, and n-3 polyunsaturated FAs (PUFAs) were significantly enriched in Mitogen-Activated Protein Kinase (MAPK) signaling, in agreement with previous studies suggesting that several MAPK players may have a primary role in regulating lipid deposition. These results give an insight into the molecular cascade associated with different IMF FA composition in ILW heavy pigs. Further studies are needed to validate the results and confirm whether some of the identified key genes may be effective candidates for pork quality.
Keywords: RNA sequencing; WGCNA; co-expression network; fat; intramuscular fat; mRNA; meat; muscle acidic composition; pork; swine.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
Genes Related to Fat Metabolism in Pigs and Intramuscular Fat Content of Pork: A Focus on Nutrigenetics and Nutrigenomics.Animals (Basel). 2022 Jan 8;12(2):150. doi: 10.3390/ani12020150. Animals (Basel). 2022. PMID: 35049772 Free PMC article. Review.
-
Muscle transcriptome analysis identifies genes involved in ciliogenesis and the molecular cascade associated with intramuscular fat content in Large White heavy pigs.PLoS One. 2020 May 19;15(5):e0233372. doi: 10.1371/journal.pone.0233372. eCollection 2020. PLoS One. 2020. PMID: 32428048 Free PMC article.
-
Multi-omics integration reveals the regulatory mechanisms of APC and CREB5 genes in lipid biosynthesis and fatty acid composition in pigs.Food Chem. 2025 Aug 1;482:143999. doi: 10.1016/j.foodchem.2025.143999. Epub 2025 Mar 22. Food Chem. 2025. PMID: 40187300
-
Proteomic and lipidomic analyses reveal saturated fatty acids, phosphatidylinositol, phosphatidylserine, and associated proteins contributing to intramuscular fat deposition.J Proteomics. 2021 Jun 15;241:104235. doi: 10.1016/j.jprot.2021.104235. Epub 2021 Apr 22. J Proteomics. 2021. PMID: 33894376
-
TRIENNIAL GROWTH AND DEVELOPMENT SYMPOSIUM: Molecular mechanisms related to bovine intramuscular fat deposition in the longissimus muscle.J Anim Sci. 2017 May;95(5):2284-2303. doi: 10.2527/jas.2016.1160. J Anim Sci. 2017. PMID: 28727015 Review.
Cited by
-
Using Machine Learning to Identify Biomarkers Affecting Fat Deposition in Pigs by Integrating Multisource Transcriptome Information.J Agric Food Chem. 2022 Aug 24;70(33):10359-10370. doi: 10.1021/acs.jafc.2c03339. Epub 2022 Aug 11. J Agric Food Chem. 2022. PMID: 35953074 Free PMC article.
-
Genes Related to Fat Metabolism in Pigs and Intramuscular Fat Content of Pork: A Focus on Nutrigenetics and Nutrigenomics.Animals (Basel). 2022 Jan 8;12(2):150. doi: 10.3390/ani12020150. Animals (Basel). 2022. PMID: 35049772 Free PMC article. Review.
-
Identification of Potential Sex-Specific Biomarkers in Pigs with Low and High Intramuscular Fat Content Using Integrated Bioinformatics and Machine Learning.Genes (Basel). 2023 Aug 25;14(9):1695. doi: 10.3390/genes14091695. Genes (Basel). 2023. PMID: 37761835 Free PMC article.
-
Molecular Pathways and Key Genes Associated With Breast Width and Protein Content in White Striping and Wooden Breast Chicken Pectoral Muscle.Front Physiol. 2022 Jul 8;13:936768. doi: 10.3389/fphys.2022.936768. eCollection 2022. Front Physiol. 2022. PMID: 35874513 Free PMC article.
References
-
- OECD . OECD-FAO Agricultural Outlook 2016-2025. OECD Publishing; Paris, France: 2016. Meat. - DOI
-
- Dalle Zotte A., Brugiapaglia A., Cullere M. What Is Meat in Italy? Anim. Front. 2017;7:63–70. doi: 10.2527/af.2017.0448. - DOI
-
- Ismea Mercati—Carni—Carne Suina e Salumi. [(accessed on 11 November 2020)]; Available online: http://www.ismeamercati.it/carni/carne-suina-salumi.
-
- Bosi P., Russo V. The Production of the Heavy Pig for High Quality Processed Products. Ital. J. Anim. Sci. 2004;3:309–321. doi: 10.4081/ijas.2004.309. - DOI
-
- Fiego D.P.L., Macchioni P., Minelli G., Santoro P. Lipid Composition of Covering and Intramuscular Fat in Pigs at Different Slaughter Age. Ital. J. Anim. Sci. 2010;9:e39. doi: 10.4081/ijas.2010.e39. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

