The Alphaviral Capsid Protein Inhibits IRAK1-Dependent TLR Signaling
- PMID: 33673546
- PMCID: PMC7997285
- DOI: 10.3390/v13030377
The Alphaviral Capsid Protein Inhibits IRAK1-Dependent TLR Signaling
Abstract
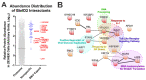
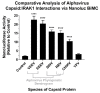
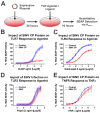
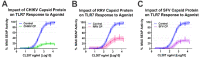
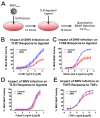
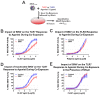
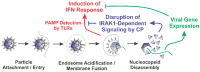
Alphaviruses are arthropod-borne RNA viruses which can cause either mild to severe febrile arthritis which may persist for months, or encephalitis which can lead to death or lifelong cognitive impairments. The non-assembly molecular role(s), functions, and protein-protein interactions of the alphavirus capsid proteins have been largely overlooked. Here we detail the use of a BioID2 biotin ligase system to identify the protein-protein interactions of the Sindbis virus capsid protein. These efforts led to the discovery of a series of novel host-pathogen interactions, including the identification of an interaction between the alphaviral capsid protein and the host IRAK1 protein. Importantly, this capsid-IRAK1 interaction is conserved across multiple alphavirus species, including arthritogenic alphaviruses SINV, Ross River virus, and Chikungunya virus; and encephalitic alphaviruses Eastern Equine Encephalitis virus, and Venezuelan Equine Encephalitis virus. The impact of the capsid-IRAK1 interaction was evaluated using a robust set of cellular model systems, leading to the realization that the alphaviral capsid protein specifically inhibits IRAK1-dependent signaling. This inhibition represents a means by which alphaviruses may evade innate immune detection and activation prior to viral gene expression. Altogether, these data identify novel capsid protein-protein interactions, establish the capsid-IRAK1 interaction as a common alphavirus host-pathogen interface, and delineate the molecular consequences of the capsid-IRAK1 interaction on IRAK1-dependent signaling.
Keywords: IRAK1; alphavirus; capsid; toll like receptors (TLR).
Conflict of interest statement
The authors declare no conflict of interest. The funding agencies had no role in the study design, data collection and analysis, decision to publish, or the preparation of the manuscript.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

