Artificial Intelligence to Get Insights of Multi-Drug Resistance Risk Factors during the First 48 Hours from ICU Admission
- PMID: 33673564
- PMCID: PMC7997208
- DOI: 10.3390/antibiotics10030239
Artificial Intelligence to Get Insights of Multi-Drug Resistance Risk Factors during the First 48 Hours from ICU Admission
Abstract
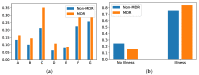
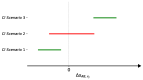
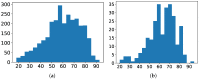
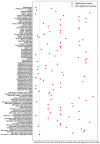
Multi-drug resistance (MDR) is one of the most current and greatest threats to the global health system nowadays. This situation is especially relevant in Intensive Care Units (ICUs), where the critical health status of these patients makes them more vulnerable. Since MDR confirmation by the microbiology laboratory usually takes 48 h, we propose several artificial intelligence approaches to get insights of MDR risk factors during the first 48 h from the ICU admission. We considered clinical and demographic features, mechanical ventilation and the antibiotics taken by the patients during this time interval. Three feature selection strategies were applied to identify statistically significant differences between MDR and non-MDR patient episodes, ending up in 24 selected features. Among them, SAPS III and Apache II scores, the age and the department of origin were identified. Considering these features, we analyzed the potential of machine learning methods for predicting whether a patient will develop a MDR germ during the first 48 h from the ICU admission. Though the results presented here are just a first incursion into this problem, artificial intelligence approaches have a great impact in this scenario, especially when enriching the set of features from the electronic health records.
Keywords: Intensive Care Unit; antibiotics; artificial intelligence; feature selection; machine learning; multi-drug resistance; risk factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- De la Bédoyère G. The Discovery of Penicillin. Evans Brothers Ltd.; London, UK: 2005.
-
- Franklin T.J., Snow G.A. Biochemistry of Antimicrobial Action. Springer; Berlin/Heidelberg, Germany: 2013.
-
- Béahdy J. Recent developments of antibiotic research and classification of antibiotics according to chemical structure. Adv. Appl. Microbiol. 1974;18:309–406. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

