The Functional Role of Loops and Flanking Sequences of G-Quadruplex Aptamer to the Hemagglutinin of Influenza a Virus
- PMID: 33673708
- PMCID: PMC7957560
- DOI: 10.3390/ijms22052409
The Functional Role of Loops and Flanking Sequences of G-Quadruplex Aptamer to the Hemagglutinin of Influenza a Virus
Abstract
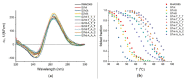
Nucleic acid aptamers are generally accepted as promising elements for the specific and high-affinity binding of various biomolecules. It has been shown for a number of aptamers that the complexes with several related proteins may possess a similar affinity. An outstanding example is the G-quadruplex DNA aptamer RHA0385, which binds to the hemagglutinins of various influenza A virus strains. These hemagglutinins have homologous tertiary structures but moderate-to-low amino acid sequence identities. Here, the experiment was inverted, targeting the same protein using a set of related, parallel G-quadruplexes. The 5'- and 3'-flanking sequences of RHA0385 were truncated to yield parallel G-quadruplex with three propeller loops that were 7, 1, and 1 nucleotides in length. Next, a set of minimal, parallel G-quadruplexes with three single-nucleotide loops was tested. These G-quadruplexes were characterized both structurally and functionally. All parallel G-quadruplexes had affinities for both recombinant hemagglutinin and influenza virions. In summary, the parallel G-quadruplex represents a minimal core structure with functional activity that binds influenza A hemagglutinin. The flanking sequences and loops represent additional features that can be used to modulate the affinity. Thus, the RHA0385-hemagglutinin complex serves as an excellent example of the hypothesis of a core structure that is decorated with additional recognizing elements capable of improving the binding properties of the aptamer.
Keywords: DNA aptamer; G-quadruplex; affinity; hemagglutinin; influenza virus; structure−activity relationship.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Novoseltseva A., Zavyalova E., Golovin A., Kopylov A. An insight into aptamer–protein complexes. Aptamers. 2018;2:1–19.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

