Diagnostic yield of genetic testing in a heterogeneous cohort of 1376 HCM patients
- PMID: 33673806
- PMCID: PMC7934228
- DOI: 10.1186/s12872-021-01927-5
Diagnostic yield of genetic testing in a heterogeneous cohort of 1376 HCM patients
Abstract
Background: Genetic testing in hypertrophic cardiomyopathy (HCM) is a published guideline-based recommendation. The diagnostic yield of genetic testing and corresponding HCM-associated genes have been largely documented by single center studies and carefully selected patient cohorts. Our goal was to evaluate the diagnostic yield of genetic testing in a heterogeneous cohort of patients with a clinical suspicion of HCM, referred for genetic testing from multiple centers around the world.
Methods: A retrospective review of patients with a suspected clinical diagnosis of HCM referred for genetic testing at Blueprint Genetics was undertaken. The analysis included syndromic, myopathic and metabolic etiologies. Genetic test results and variant classifications were extracted from the database. Variants classified as pathogenic (P) or likely pathogenic (LP) were considered diagnostic.
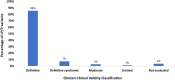
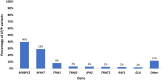
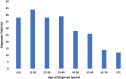
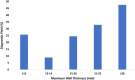
Results: A total of 1376 samples were analyzed. Three hundred and sixty-nine tests were diagnostic (26.8%); 373 P or LP variants were identified. Only one copy number variant was identified. The majority of diagnostic variants involved genes encoding the sarcomere (85.0%) followed by 4.3% of diagnostic variants identified in the RASopathy genes. Two percent of diagnostic variants were in genes associated with a cardiomyopathy other than HCM or an inherited arrhythmia. Clinical variables that increased the likelihood of identifying a diagnostic variant included: an earlier age at diagnosis (p < 0.0001), a higher maximum wall thickness (MWT) (p < 0.0001), a positive family history (p < 0.0001), the absence of hypertension (p = 0.0002), and the presence of an implantable cardioverter-defibrillator (ICD) (p = 0.0004).
Conclusion: The diagnostic yield of genetic testing in this heterogeneous cohort of patients with a clinical suspicion of HCM is lower than what has been reported in well-characterized patient cohorts. We report the highest yield of diagnostic variants in the RASopathy genes identified in a laboratory cohort of HCM patients to date. The spectrum of genes implicated in this unselected cohort highlights the importance of pre-and post-test counseling when offering genetic testing to the broad HCM population.
Keywords: Counseling; Diagnosis; Genetic testing; Hypertrophic cardiomyopathy; Next generation sequencing.
Conflict of interest statement
Minor conflict of interest: SM, TPA, JK are co-founders of Blueprint Genetics and JH, IS, JT, EHS, ST, TKK, JS, JTo, VK, MV, MM, JSi, MG, PS, SM, JP, TPA, and JK are full-time employees of Blueprint Genetics, which offers genetic diagnostic services.
Figures
References
-
- Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, Charron P, et al. 2014 ESC guidelines on diagnosis and management of hypertrophic cardiomyopathythe task force for the diagnosis and management of hypertrophic cardiomyopathy of the european society of cardiology (ESC) Eur Heart J. 2014;35(39):2733–2779. doi: 10.1093/eurheartj/ehu284. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

