Rapid Detection and Signaling of DNA Damage by PARP-1
- PMID: 33674152
- PMCID: PMC8364484
- DOI: 10.1016/j.tibs.2021.01.014
Rapid Detection and Signaling of DNA Damage by PARP-1
Abstract
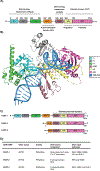
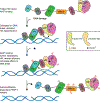
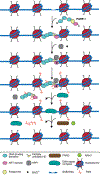
Poly(ADP-ribosyl) polymerase-1 (PARP-1) is an abundant ADP-ribosyl transferase that regulates various biological processes. PARP-1 is widely recognized as a first-line responder molecule in DNA damage response (DDR). Here, we review the full cycle of detecting DNA damage by PARP-1, PARP-1 activation upon DNA binding, and PARP-1 release from a DNA break. We also discuss the allosteric consequence upon binding of PARP inhibitors (PARPi) and the opportunity to tune its release from a DNA break. It is now possible to harness this new understanding to design novel PARPi for treating diseases where cell toxicity caused by PARP-1 'trapping' on DNA is either the desired consequence or entirely counterproductive.
Keywords: DNA damage response; PARPi; PARylation; poly(ADP-ribose) glycohydrolase.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Interests B.E.B. is a founder of Hysplex LLC, with interests in PARPi development.
Figures
References
-
- Gibson BA and Kraus WL (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell. Biol 13, 411–424 - PubMed
-
- Amé JC et al. (2004) The PARP superfamily. Bioessays 26, 882–893 - PubMed
-
- Chambon JD et al. (1963) Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun 11, 39–43 - PubMed
-
- Amé JC et al. (1999) PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem 274, 17860–17868 - PubMed
-
- Rulten SL et al. (2011) PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell 41, 33–45 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

